当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Order, Disorder, and Temperature-Driven Compaction in a Designed Elastin Protein
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-02-20 00:00:00 , DOI: 10.1021/acs.jpcb.7b11596 Kelly N. Greenland 1 , Ma. Faye Charmagne A. Carvajal 2 , Jonathan M. Preston 1 , Siri Ekblad 1 , William L. Dean 3 , Jeff Y. Chiang 1 , Ronald L. Koder 1, 4 , Richard J. Wittebort 2
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-02-20 00:00:00 , DOI: 10.1021/acs.jpcb.7b11596 Kelly N. Greenland 1 , Ma. Faye Charmagne A. Carvajal 2 , Jonathan M. Preston 1 , Siri Ekblad 1 , William L. Dean 3 , Jeff Y. Chiang 1 , Ronald L. Koder 1, 4 , Richard J. Wittebort 2
Affiliation
|
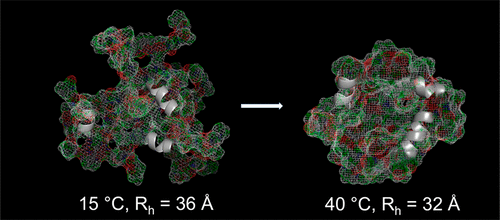
Artificial minielastin constructs have been designed that replicate the structure and function of natural elastins in a simpler context, allowing the NMR observation of structure and dynamics of elastin-like proteins with complete residue-specific resolution. We find that the alanine-rich cross-linking domains of elastin have a partially helical structure, but only when capped by proline-rich hydrophobic domains. We also find that the hydrophobic domains, composed of prominent 6-residue repeats VPGVGG and APGVGV found in natural elastins, appear random coil by both NMR chemical shift analysis and circular dichroism. However, these elastin hydrophobic domains exhibit structural bias for a dynamically disordered conformation that is neither helical nor β sheet with a degree of nonrandom structural bias which is dependent on residue type and position in the sequence. Another nonrandom-coil aspect of hydrophobic domain structure lies in the fact that, in contrast to other intrinsically disordered proteins, these hydrophobic domains retain a relatively condensed conformation whether attached to cross-linking domains or not. Importantly, these domains and the proteins containing them constrict with increasing temperature by up to 30% in volume without becoming more ordered. This property is often observed in nonbiological polymers and suggests that temperature-driven constriction is a new type of protein structural change that is linked to elastin’s biological functions of coacervation-driven assembly and elastic recoil.
中文翻译:

设计的弹性蛋白中的有序,无序和温度驱动的压实
已设计了人工小弹性蛋白构建体,该结构可在更简单的环境中复制天然弹性蛋白的结构和功能,从而使NMR可以观察到具有完整残基特异性分辨率的弹性蛋白样蛋白的结构和动力学。我们发现,弹性蛋白的富含丙氨酸的交联结构域具有部分螺旋结构,但仅当被富含脯氨酸的疏水性结构域覆盖时才如此。我们还发现,由天然弹性蛋白中发现的突出的6个残基重复序列VPGVGG和APGVGV组成的疏水域,通过NMR化学位移分析和圆二色性显示为无规卷曲。然而,这些弹性蛋白疏水结构域表现出对于既不是螺旋结构也不是β折叠的动态无序构象的结构偏倚,其非随机结构偏倚的程度取决于残基的类型和序列中的位置。疏水域结构的另一个非随机螺旋方面在于,与其他内在无序的蛋白质相反,这些疏水域无论是否连接到交联域都保留相对缩合的构象。重要的是,这些结构域和包含它们的蛋白质会随着温度的升高而收缩,体积最多增加30%,而不会变得更有序。
更新日期:2018-02-20
中文翻译:

设计的弹性蛋白中的有序,无序和温度驱动的压实
已设计了人工小弹性蛋白构建体,该结构可在更简单的环境中复制天然弹性蛋白的结构和功能,从而使NMR可以观察到具有完整残基特异性分辨率的弹性蛋白样蛋白的结构和动力学。我们发现,弹性蛋白的富含丙氨酸的交联结构域具有部分螺旋结构,但仅当被富含脯氨酸的疏水性结构域覆盖时才如此。我们还发现,由天然弹性蛋白中发现的突出的6个残基重复序列VPGVGG和APGVGV组成的疏水域,通过NMR化学位移分析和圆二色性显示为无规卷曲。然而,这些弹性蛋白疏水结构域表现出对于既不是螺旋结构也不是β折叠的动态无序构象的结构偏倚,其非随机结构偏倚的程度取决于残基的类型和序列中的位置。疏水域结构的另一个非随机螺旋方面在于,与其他内在无序的蛋白质相反,这些疏水域无论是否连接到交联域都保留相对缩合的构象。重要的是,这些结构域和包含它们的蛋白质会随着温度的升高而收缩,体积最多增加30%,而不会变得更有序。











































 京公网安备 11010802027423号
京公网安备 11010802027423号