当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Towards enhanced metabolomic data analysis of mass spectrometry image: Multivariate Curve Resolution and machine learning
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2018-12-01 , DOI: 10.1016/j.aca.2018.02.031 Xiang Tian , Genwei Zhang , Yihan Shao , Zhibo Yang
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2018-12-01 , DOI: 10.1016/j.aca.2018.02.031 Xiang Tian , Genwei Zhang , Yihan Shao , Zhibo Yang
|
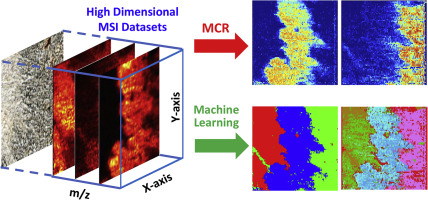
Large amounts of data are generally produced from mass spectrometry imaging (MSI) experiments in obtaining the molecular and spatial information of biological samples. Traditionally, MS images are constructed using manually selected ions, and it is very challenging to comprehensively analyze MSI results due to their large data sizes and highly complex data structures. To overcome these barriers, it is obligatory to develop advanced data analysis approaches to handle the increasingly large MSI data. In the current study, we focused on the method development of using Multivariate Curve Resolution (MCR) and Machine Learning (ML) approaches. We aimed to effectively extract the essential information present in the large and complex MSI data and enhance the metabolomic data analysis of biological tissues. Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS) algorithm was used to obtain major patterns of spatial distribution and grouped metabolites with the same spatial distribution patterns. In addition, both supervised and unsupervised ML methods were established to analyze the MSI data. In the supervised ML approach, Random Forest method was selected, and the model was trained using the selected datasets based on the distribution pattern obtained from MCR-ALS analyses. In the unsupervised ML approach, both DBSCAN (Density-based Spatial Clustering of Applications with Noise) and CLARA (Clustering Large Applications) were applied to cluster the MSI datasets. It is worth noting that similar patterns of spatial distribution were discovered through MSI data analysis using MCR-ALS, supervised ML, and unsupervised ML. Our protocols of data analysis can be applied to process the data acquired using many other types of MSI techniques, and to extract the overall features present in MSI results that are intractable using traditional data analysis approaches.
中文翻译:

质谱图像的增强代谢组学数据分析:多元曲线分辨率和机器学习
在获取生物样品的分子和空间信息时,通常从质谱成像 (MSI) 实验中产生大量数据。传统上,MS图像是使用手动选择的离子构建的,并且由于其数据量大且数据结构高度复杂,因此对MSI结果进行综合分析非常具有挑战性。为了克服这些障碍,必须开发先进的数据分析方法来处理越来越大的 MSI 数据。在当前的研究中,我们专注于使用多元曲线分辨率 (MCR) 和机器学习 (ML) 方法的方法开发。我们旨在有效地提取大量复杂的 MSI 数据中存在的基本信息,并增强生物组织的代谢组学数据分析。多元曲线分辨率-交替最小二乘法 (MCR-ALS) 算法用于获得空间分布的主要模式和具有相同空间分布模式的分组代谢物。此外,建立了有监督和无监督的 ML 方法来分析 MSI 数据。在有监督的 ML 方法中,选择了随机森林方法,并根据从 MCR-ALS 分析中获得的分布模式,使用选定的数据集训练模型。在无监督 ML 方法中,DBSCAN(基于密度的应用程序空间聚类与噪声)和 CLARA(大型应用程序聚类)都被应用于对 MSI 数据集进行聚类。值得注意的是,通过使用 MCR-ALS、监督 ML 和无监督 ML 的 MSI 数据分析发现了类似的空间分布模式。
更新日期:2018-12-01
中文翻译:

质谱图像的增强代谢组学数据分析:多元曲线分辨率和机器学习
在获取生物样品的分子和空间信息时,通常从质谱成像 (MSI) 实验中产生大量数据。传统上,MS图像是使用手动选择的离子构建的,并且由于其数据量大且数据结构高度复杂,因此对MSI结果进行综合分析非常具有挑战性。为了克服这些障碍,必须开发先进的数据分析方法来处理越来越大的 MSI 数据。在当前的研究中,我们专注于使用多元曲线分辨率 (MCR) 和机器学习 (ML) 方法的方法开发。我们旨在有效地提取大量复杂的 MSI 数据中存在的基本信息,并增强生物组织的代谢组学数据分析。多元曲线分辨率-交替最小二乘法 (MCR-ALS) 算法用于获得空间分布的主要模式和具有相同空间分布模式的分组代谢物。此外,建立了有监督和无监督的 ML 方法来分析 MSI 数据。在有监督的 ML 方法中,选择了随机森林方法,并根据从 MCR-ALS 分析中获得的分布模式,使用选定的数据集训练模型。在无监督 ML 方法中,DBSCAN(基于密度的应用程序空间聚类与噪声)和 CLARA(大型应用程序聚类)都被应用于对 MSI 数据集进行聚类。值得注意的是,通过使用 MCR-ALS、监督 ML 和无监督 ML 的 MSI 数据分析发现了类似的空间分布模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号