当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
AMOEBA Polarizable Atomic Multipole Force Field for Nucleic Acids
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-02-13 00:00:00 , DOI: 10.1021/acs.jctc.7b01169 Changsheng Zhang 1 , Chao Lu 2 , Zhifeng Jing 1 , Chuanjie Wu 2 , Jean-Philip Piquemal 1, 3 , Jay W Ponder 2 , Pengyu Ren 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-02-13 00:00:00 , DOI: 10.1021/acs.jctc.7b01169 Changsheng Zhang 1 , Chao Lu 2 , Zhifeng Jing 1 , Chuanjie Wu 2 , Jean-Philip Piquemal 1, 3 , Jay W Ponder 2 , Pengyu Ren 1
Affiliation
|
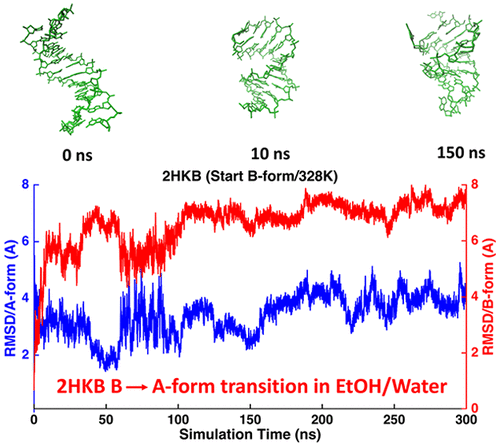
The AMOEBA polarizable atomic multipole force field for nucleic acids is presented. Valence and electrostatic parameters were determined from high-level quantum mechanical data, including structures, conformational energy, and electrostatic potentials, of nucleotide model compounds. Previously derived parameters for the phosphate group and nucleobases were incorporated. A total of over 35 μs of condensed-phase molecular dynamics simulations of DNA and RNA molecules in aqueous solution and crystal lattice were performed to validate and refine the force field. The solution and/or crystal structures of DNA B-form duplexes, RNA duplexes, and hairpins were captured with an average root-mean-squared deviation from NMR structures below or around 2.0 Å. Structural details, such as base pairing and stacking, sugar puckering, backbone and χ-torsion angles, groove geometries, and crystal packing interfaces, agreed well with NMR and/or X-ray. The interconversion between A- and B-form DNAs was observed in ethanol–water mixtures at 328 K. Crystal lattices of B- and Z-form DNA and A-form RNA were examined with simulations. For the RNA tetraloop, single strand tetramers, and HIV TAR with 29 residues, the simulated conformational states, 3J-coupling, nuclear Overhauser effect, and residual dipolar coupling data were compared with NMR results. Starting from a totally unstacked/unfolding state, the rCAAU tetranucleotide was folded into A-form-like structures during ∼1 μs molecular dynamics simulations.
中文翻译:

AMOEBA 核酸可极化原子多极力场
提出了核酸的 AMOEBA 极化原子多极力场。化合价和静电参数是根据高级量子力学数据确定的,包括核苷酸模型化合物的结构、构象能和静电势。先前导出的磷酸基团和核碱基参数被纳入。对水溶液和晶格中的 DNA 和 RNA 分子进行了总计超过 35 μs 的凝聚相分子动力学模拟,以验证和细化力场。捕获的 DNA B 型双链体、RNA 双链体和发夹的溶液和/或晶体结构与 NMR 结构的平均均方根偏差低于或约为 2.0 Å。结构细节,例如碱基配对和堆叠、糖褶皱、骨架和 χ 扭转角、凹槽几何形状和晶体堆积界面,与 NMR 和/或 X 射线非常吻合。在 328 K 的乙醇-水混合物中观察到 A 型和 B 型 DNA 之间的相互转化。通过模拟检查了 B 型和 Z 型 DNA 以及 A 型 RNA 的晶格。对于RNA四环、单链四聚体和具有29个残基的HIV TAR,将模拟的构象状态、 3J耦合、核奥弗豪瑟效应和残余偶极耦合数据与NMR结果进行比较。从完全未堆叠/展开状态开始,rCAAU 四核苷酸在~1 μs 分子动力学模拟过程中被折叠成 A 型结构。
更新日期:2018-02-13
中文翻译:

AMOEBA 核酸可极化原子多极力场
提出了核酸的 AMOEBA 极化原子多极力场。化合价和静电参数是根据高级量子力学数据确定的,包括核苷酸模型化合物的结构、构象能和静电势。先前导出的磷酸基团和核碱基参数被纳入。对水溶液和晶格中的 DNA 和 RNA 分子进行了总计超过 35 μs 的凝聚相分子动力学模拟,以验证和细化力场。捕获的 DNA B 型双链体、RNA 双链体和发夹的溶液和/或晶体结构与 NMR 结构的平均均方根偏差低于或约为 2.0 Å。结构细节,例如碱基配对和堆叠、糖褶皱、骨架和 χ 扭转角、凹槽几何形状和晶体堆积界面,与 NMR 和/或 X 射线非常吻合。在 328 K 的乙醇-水混合物中观察到 A 型和 B 型 DNA 之间的相互转化。通过模拟检查了 B 型和 Z 型 DNA 以及 A 型 RNA 的晶格。对于RNA四环、单链四聚体和具有29个残基的HIV TAR,将模拟的构象状态、 3J耦合、核奥弗豪瑟效应和残余偶极耦合数据与NMR结果进行比较。从完全未堆叠/展开状态开始,rCAAU 四核苷酸在~1 μs 分子动力学模拟过程中被折叠成 A 型结构。










































 京公网安备 11010802027423号
京公网安备 11010802027423号