当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Interactive Visual Exploration of 3D Mass Spectrometry Imaging Data Using Hierarchical Stochastic Neighbor Embedding Reveals Spatiomolecular Structures at Full Data Resolution.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-02-15 , DOI: 10.1021/acs.jproteome.7b00725 Walid M Abdelmoula 1, 2 , Nicola Pezzotti 3 , Thomas Hölt 3 , Jouke Dijkstra 1 , Anna Vilanova 3 , Liam A McDonnell 4 , Boudewijn P F Lelieveldt 1, 3
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-02-15 , DOI: 10.1021/acs.jproteome.7b00725 Walid M Abdelmoula 1, 2 , Nicola Pezzotti 3 , Thomas Hölt 3 , Jouke Dijkstra 1 , Anna Vilanova 3 , Liam A McDonnell 4 , Boudewijn P F Lelieveldt 1, 3
Affiliation
|
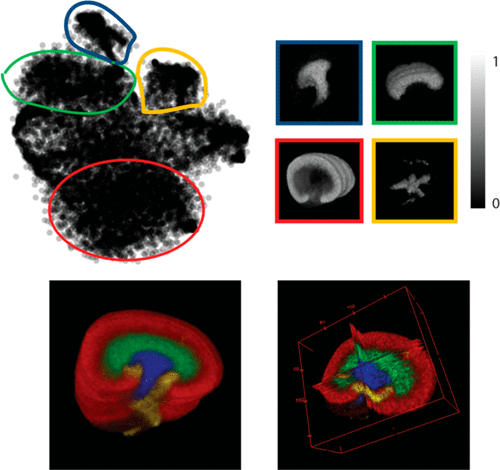
Technological advances in mass spectrometry imaging (MSI) have contributed to growing interest in 3D MSI. However, the large size of 3D MSI data sets has made their efficient analysis and visualization and the identification of informative molecular patterns computationally challenging. Hierarchical stochastic neighbor embedding (HSNE), a nonlinear dimensionality reduction technique that aims at finding hierarchical and multiscale representations of large data sets, is a recent development that enables the analysis of millions of data points, with manageable time and memory complexities. We demonstrate that HSNE can be used to analyze large 3D MSI data sets at full mass spectral and spatial resolution. To benchmark the technique as well as demonstrate its broad applicability, we have analyzed a number of publicly available 3D MSI data sets, recorded from various biological systems and spanning different mass-spectrometry ionization techniques. We demonstrate that HSNE is able to rapidly identify regions of interest within these large high-dimensionality data sets as well as aid the identification of molecular ions that characterize these regions of interest; furthermore, through clearly separating measurement artifacts, the HSNE analysis exhibits a degree of robustness to measurement batch effects, spatially correlated noise, and mass spectral misalignment.
中文翻译:

使用分层随机邻居嵌入技术对3D质谱成像数据进行交互式视觉探索,可以完整的数据分辨率显示空间分子结构。
质谱成像(MSI)的技术进步已引起人们对3D MSI的日益增长的兴趣。但是,庞大的3D MSI数据集使其高效的分析和可视化以及信息分子模式的识别在计算上具有挑战性。分层随机邻居嵌入(HSNE)是旨在减少大型数据集的分层和多尺度表示的一种非线性降维技术,它是最近的一项发展,它能够分析数百万个数据点,并具有可管理的时间和存储复杂性。我们证明,HSNE可用于以完整的质谱和空间分辨率分析大型3D MSI数据集。为了对这项技术进行基准测试并证明其广泛的适用性,我们分析了许多公开可用的3D MSI数据集,记录了来自各种生物系统的信息,并涵盖了不同的质谱电离技术。我们证明,HSNE能够快速识别这些大型高维数据集中的目标区域,并有助于识别表征这些目标区域的分子离子;此外,通过清楚地分离测量伪影,HSNE分析对测量批次效应,空间相关噪声和质谱未对准表现出一定程度的鲁棒性。
更新日期:2018-02-16
中文翻译:

使用分层随机邻居嵌入技术对3D质谱成像数据进行交互式视觉探索,可以完整的数据分辨率显示空间分子结构。
质谱成像(MSI)的技术进步已引起人们对3D MSI的日益增长的兴趣。但是,庞大的3D MSI数据集使其高效的分析和可视化以及信息分子模式的识别在计算上具有挑战性。分层随机邻居嵌入(HSNE)是旨在减少大型数据集的分层和多尺度表示的一种非线性降维技术,它是最近的一项发展,它能够分析数百万个数据点,并具有可管理的时间和存储复杂性。我们证明,HSNE可用于以完整的质谱和空间分辨率分析大型3D MSI数据集。为了对这项技术进行基准测试并证明其广泛的适用性,我们分析了许多公开可用的3D MSI数据集,记录了来自各种生物系统的信息,并涵盖了不同的质谱电离技术。我们证明,HSNE能够快速识别这些大型高维数据集中的目标区域,并有助于识别表征这些目标区域的分子离子;此外,通过清楚地分离测量伪影,HSNE分析对测量批次效应,空间相关噪声和质谱未对准表现出一定程度的鲁棒性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号