当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Study of the affinity between the protein kinase PKA and homoarginine-containing peptides derived from kemptide: Free energy perturbation (FEP) calculations
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2018-02-05 , DOI: 10.1002/jcc.25176 Karel Mena-Ulecia 1 , Fabian Gonzalez-Norambuena 2 , Ariela Vergara-Jaque 2 , Horacio Poblete 2 , William Tiznado 1 , Julio Caballero 2
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2018-02-05 , DOI: 10.1002/jcc.25176 Karel Mena-Ulecia 1 , Fabian Gonzalez-Norambuena 2 , Ariela Vergara-Jaque 2 , Horacio Poblete 2 , William Tiznado 1 , Julio Caballero 2
Affiliation
|
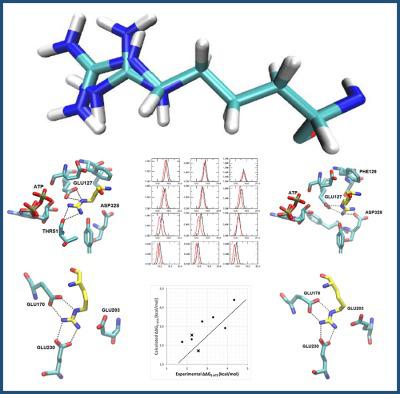
Protein kinases (PKs) discriminate between closely related sequences that contain serine, threonine, and/or tyrosine residues. Such specificity is defined by the amino acid sequence surrounding the phosphorylatable residue, so that it is possible to identify an optimal recognition motif (ORM) for each PK. The ORM for the protein kinase A (PKA), a well‐known member of the PK family, is the sequence RRX(S/T)X, where arginines at the −3 and −2 positions play a key role with respect to the primed phosphorylation site. In this work, differential affinities of PKA for the peptide substrate Kemptide (LRRASLG) and mutants that substitute the arginine residues by the unnatural peptide homoarginine were evaluated through molecular dynamics (MD) and free energy perturbation (FEP) calculations. The FEP study for the homoarginine mutants required previous elaboration of a CHARMM “arginine to homoarginine” (R2B) hybrid topology file which is available in this manuscript as Supporting Information. Mutants substituting the arginine residues by alanine, lysine, and histidine were also considered in the comparison by using the same protocol. FEP calculations allowed estimating the free energy changes from the free PKA to PKA‐substrate complex (ΔΔGE→ES) when Kemptide structure was mutated. Both ΔΔGS→ES values for homoarginine mutants were predicted with a difference below 1 kcal/mol. In addition, FEP correctly predicted that all the studied mutations decrease the catalytic efficiency of Kemptide for PKA. © 2018 Wiley Periodicals, Inc.
中文翻译:

研究蛋白激酶 PKA 与源自 kemptide 的高精氨酸肽之间的亲和力:自由能扰动 (FEP) 计算
蛋白激酶 (PK) 区分包含丝氨酸、苏氨酸和/或酪氨酸残基的密切相关序列。这种特异性由可磷酸化残基周围的氨基酸序列定义,因此可以识别每个 PK 的最佳识别基序 (ORM)。蛋白激酶 A (PKA) 是 PK 家族的知名成员,其 ORM 是序列 RRX(S/T)X,其中 -3 和 -2 位的精氨酸在引发磷酸化位点。在这项工作中,通过分子动力学 (MD) 和自由能扰动 (FEP) 计算评估了 PKA 对肽底物 Kemptide (LRRASLG) 和用非天然肽高精氨酸替代精氨酸残基的突变体的差异亲和力。高精氨酸突变体的 FEP 研究需要事先详细阐述 CHARMM“精氨酸到高精氨酸”(R2B)混合拓扑文件,该文件在本手稿中作为支持信息提供。通过使用相同的方案,在比较中还考虑了用丙氨酸、赖氨酸和组氨酸取代精氨酸残基的突变体。当 Kemptide 结构发生突变时,FEP 计算允许估计从游离 PKA 到 PKA 底物复合物(ΔΔGE→ES)的自由能变化。高精氨酸突变体的两个 ΔΔGS→ES 值均以低于 1 kcal/mol 的差异进行预测。此外,FEP 正确预测所有研究的突变都会降低 Kemptide 对 PKA 的催化效率。© 2018 Wiley Periodicals, Inc.
更新日期:2018-02-05
中文翻译:

研究蛋白激酶 PKA 与源自 kemptide 的高精氨酸肽之间的亲和力:自由能扰动 (FEP) 计算
蛋白激酶 (PK) 区分包含丝氨酸、苏氨酸和/或酪氨酸残基的密切相关序列。这种特异性由可磷酸化残基周围的氨基酸序列定义,因此可以识别每个 PK 的最佳识别基序 (ORM)。蛋白激酶 A (PKA) 是 PK 家族的知名成员,其 ORM 是序列 RRX(S/T)X,其中 -3 和 -2 位的精氨酸在引发磷酸化位点。在这项工作中,通过分子动力学 (MD) 和自由能扰动 (FEP) 计算评估了 PKA 对肽底物 Kemptide (LRRASLG) 和用非天然肽高精氨酸替代精氨酸残基的突变体的差异亲和力。高精氨酸突变体的 FEP 研究需要事先详细阐述 CHARMM“精氨酸到高精氨酸”(R2B)混合拓扑文件,该文件在本手稿中作为支持信息提供。通过使用相同的方案,在比较中还考虑了用丙氨酸、赖氨酸和组氨酸取代精氨酸残基的突变体。当 Kemptide 结构发生突变时,FEP 计算允许估计从游离 PKA 到 PKA 底物复合物(ΔΔGE→ES)的自由能变化。高精氨酸突变体的两个 ΔΔGS→ES 值均以低于 1 kcal/mol 的差异进行预测。此外,FEP 正确预测所有研究的突变都会降低 Kemptide 对 PKA 的催化效率。© 2018 Wiley Periodicals, Inc.









































 京公网安备 11010802027423号
京公网安备 11010802027423号