当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Investigating the Network Basis of Negative Genetic Interactions in Saccharomyces cerevisiae with Integrated Biological Networks and Triplet Motif Analysis
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-02-08 00:00:00 , DOI: 10.1021/acs.jproteome.7b00649 Chi Nam Ignatius Pang 1 , Apurv Goel 1 , Marc R. Wilkins 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-02-08 00:00:00 , DOI: 10.1021/acs.jproteome.7b00649 Chi Nam Ignatius Pang 1 , Apurv Goel 1 , Marc R. Wilkins 1
Affiliation
|
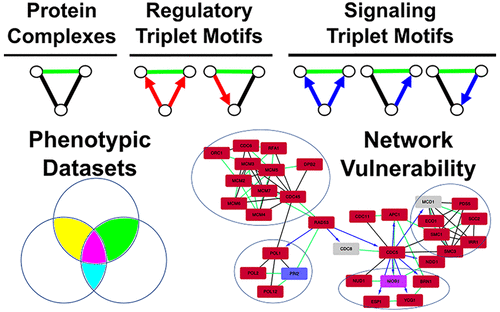
Negative genetic interactions in Saccharomyces cerevisiae have been systematically screened to near-completeness, with >500 000 interactions identified. Nevertheless, the biological basis of these interactions remains poorly understood. To investigate this, we analyzed negative genetic interactions within an integrated biological network, being the union of protein–protein, kinase–substrate, and transcription factor–target gene interactions. Network triplets, containing two genes/proteins that show negative genetic interaction and a third protein from the network, were then analyzed. Strikingly, just six out of 15 possible triplet motif types were present, as compared to randomized networks. These were in three clear groups: protein–protein interactions, signaling, and regulatory triplets where the latter two showed no overlap. In the triplets, negative genetic interactions were associated with paralogs and ohnologs; however, these were very rare. Negative genetic interactions among the six triplet motifs did however show strong dosage constraints, with genes being significantly associated with toxicity on overexpression and periodicity in the cell cycle. Negative genetic interactions overlapped with other interaction types in 37% of cases; these were predominantly associated with protein complexes or signaling events. Finally, we highlight regions of “network vulnerability” containing multiple negative genetic interactions; these could be targeted in fungal species for the regulation of cell growth.
中文翻译:

利用集成生物网络和三重态母题分析研究酿酒酵母中负遗传相互作用的网络基础
酿酒酵母中的负遗传相互作用已对系统进行了近乎完整的筛选,并确定了> 50万个互动。然而,这些相互作用的生物学基础仍然知之甚少。为了对此进行研究,我们分析了一个整合的生物网络中的负面遗传相互作用,即蛋白质-蛋白质,激酶-底物和转录因子-靶基因相互作用的结合。然后分析了三胞胎,其中包含两个显示负遗传相互作用的基因/蛋白质和来自网络的第三个蛋白质。令人惊讶的是,与随机网络相比,在15种可能的三联体基序类型中仅存在6种。它们分为三个清晰的组:蛋白质-蛋白质相互作用,信号传导和调节三联体,其中后两个没有重叠。在三胞胎中 负面的遗传相互作用与旁系同源物和同源物相关。但是,这些非常罕见。然而,六个三联体基元之间的负遗传相互作用确实显示出强烈的剂量限制,其中基因与细胞周期中过表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。基因与细胞周期过度表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。基因与细胞周期过度表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。
更新日期:2018-02-09
中文翻译:

利用集成生物网络和三重态母题分析研究酿酒酵母中负遗传相互作用的网络基础
酿酒酵母中的负遗传相互作用已对系统进行了近乎完整的筛选,并确定了> 50万个互动。然而,这些相互作用的生物学基础仍然知之甚少。为了对此进行研究,我们分析了一个整合的生物网络中的负面遗传相互作用,即蛋白质-蛋白质,激酶-底物和转录因子-靶基因相互作用的结合。然后分析了三胞胎,其中包含两个显示负遗传相互作用的基因/蛋白质和来自网络的第三个蛋白质。令人惊讶的是,与随机网络相比,在15种可能的三联体基序类型中仅存在6种。它们分为三个清晰的组:蛋白质-蛋白质相互作用,信号传导和调节三联体,其中后两个没有重叠。在三胞胎中 负面的遗传相互作用与旁系同源物和同源物相关。但是,这些非常罕见。然而,六个三联体基元之间的负遗传相互作用确实显示出强烈的剂量限制,其中基因与细胞周期中过表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。基因与细胞周期过度表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。基因与细胞周期过度表达和周期性的毒性显着相关。阴性遗传相互作用与其他相互作用类型重叠的比例为37%;这些主要与蛋白质复合物或信号事件相关。最后,我们重点介绍了包含多个负面遗传相互作用的“网络脆弱性”区域。这些可以针对真菌物种,以调节细胞的生长。











































 京公网安备 11010802027423号
京公网安备 11010802027423号