当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure-guided approach identifies a novel class of HIV-1 ribonuclease H inhibitors: binding mode insights through magnesium complexation and site-directed mutagenesis studies†
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2018-02-01 00:00:00 , DOI: 10.1039/c7md00600d Vasanthanathan Poongavanam 1 , Angela Corona 2 , Casper Steinmann 1 , Luigi Scipione 3 , Nicole Grandi 2 , Fabiana Pandolfi 3 , Roberto Di Santo 3 , Roberta Costi 3 , Francesca Esposito 2 , Enzo Tramontano 2, 4 , Jacob Kongsted 1
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2018-02-01 00:00:00 , DOI: 10.1039/c7md00600d Vasanthanathan Poongavanam 1 , Angela Corona 2 , Casper Steinmann 1 , Luigi Scipione 3 , Nicole Grandi 2 , Fabiana Pandolfi 3 , Roberto Di Santo 3 , Roberta Costi 3 , Francesca Esposito 2 , Enzo Tramontano 2, 4 , Jacob Kongsted 1
Affiliation
|
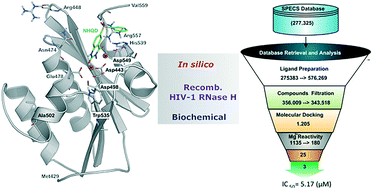
Persistent HIV infection requires lifelong treatment and among the 2.1 million new HIV infections that occur every year there is an increased rate of transmitted drug-resistant mutations. This fact requires a constant and timely effort in order to identify and develop new HIV inhibitors with innovative mechanisms. The HIV-1 reverse transcriptase (RT) associated ribonuclease H (RNase H) is the only viral encoded enzyme that still lacks an efficient inhibitor despite the fact that it is a well-validated target whose functional abrogation compromises viral infectivity. Identification of new drugs is a long and expensive process that can be speeded up by in silico methods. In the present study, a structure-guided screening is coupled with a similarity-based search on the Specs database to identify a new class of HIV-1 RNase H inhibitors. Out of the 45 compounds selected for experimental testing, 15 inhibited the RNase H function below 100 μM with three hits exhibiting IC50 values <10 μM. The most active compound, AA, inhibits HIV-1 RNase H with an IC50 of 5.1 μM and exhibits a Mg-independent mode of inhibition. Site-directed mutagenesis studies provide valuable insight into the binding mode of newly identified compounds; for instance, compound AA involves extensive interactions with a lipophilic pocket formed by Ala502, Lys503, and Trp (406, 426 and 535) and polar interactions with Arg557 and the highly conserved RNase H primer-grip residue Asn474. The structural insights obtained from this work provide the bases for further lead optimization.
中文翻译:

结构引导方法确定了一类新型 HIV-1 核糖核酸酶 H 抑制剂:通过镁络合和定点诱变研究了解结合模式†
持续性 HIV 感染需要终生治疗,每年新增 210 万例 HIV 感染,其中耐药突变传播率不断上升。这一事实需要不断、及时的努力,以识别和开发具有创新机制的新型艾滋病毒抑制剂。 HIV-1 逆转录酶 (RT) 相关核糖核酸酶 H (RNase H) 是唯一一种仍然缺乏有效抑制剂的病毒编码酶,尽管它是一个经过充分验证的靶标,其功能失效会损害病毒感染性。新药的鉴定是一个漫长而昂贵的过程,可以通过计算机方法加速。在本研究中,结构引导筛选与 Specs 数据库中基于相似性的搜索相结合,以确定一类新的 HIV-1 RNase H 抑制剂。在选择用于实验测试的 45 种化合物中,有 15 种将 RNase H 功能抑制在 100 μM 以下,其中 3 个命中表现出 IC 50值 <10 id=29>AA ,抑制 HIV-1 RNase H 的 IC 50为 5.1 μM,并表现出不依赖 Mg 的抑制模式。定点诱变研究为新发现的化合物的结合模式提供了有价值的见解;例如,化合物AA涉及与 Ala502、Lys503 和 Trp(406、426 和 535)形成的亲脂性口袋的广泛相互作用,以及与 Arg557 和高度保守的 RNase H 引物夹残基 Asn474 的极性相互作用。从这项工作中获得的结构见解为进一步先导优化提供了基础。
更新日期:2018-02-01
中文翻译:

结构引导方法确定了一类新型 HIV-1 核糖核酸酶 H 抑制剂:通过镁络合和定点诱变研究了解结合模式†
持续性 HIV 感染需要终生治疗,每年新增 210 万例 HIV 感染,其中耐药突变传播率不断上升。这一事实需要不断、及时的努力,以识别和开发具有创新机制的新型艾滋病毒抑制剂。 HIV-1 逆转录酶 (RT) 相关核糖核酸酶 H (RNase H) 是唯一一种仍然缺乏有效抑制剂的病毒编码酶,尽管它是一个经过充分验证的靶标,其功能失效会损害病毒感染性。新药的鉴定是一个漫长而昂贵的过程,可以通过计算机方法加速。在本研究中,结构引导筛选与 Specs 数据库中基于相似性的搜索相结合,以确定一类新的 HIV-1 RNase H 抑制剂。在选择用于实验测试的 45 种化合物中,有 15 种将 RNase H 功能抑制在 100 μM 以下,其中 3 个命中表现出 IC 50值 <10 id=29>AA ,抑制 HIV-1 RNase H 的 IC 50为 5.1 μM,并表现出不依赖 Mg 的抑制模式。定点诱变研究为新发现的化合物的结合模式提供了有价值的见解;例如,化合物AA涉及与 Ala502、Lys503 和 Trp(406、426 和 535)形成的亲脂性口袋的广泛相互作用,以及与 Arg557 和高度保守的 RNase H 引物夹残基 Asn474 的极性相互作用。从这项工作中获得的结构见解为进一步先导优化提供了基础。











































 京公网安备 11010802027423号
京公网安备 11010802027423号