当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-02-08 , DOI: 10.1021/acschembio.7b00890 Fei Ye 1, 2 , Xiangqian Kong 3 , Hao Zhang 2 , Yan Liu 2 , Zhiyuan Shao 2 , Jia Jin 1 , Yi Cai 3 , Rukang Zhang 2 , Linjuan Li 2 , Yang W Zhang 3 , Yu-Chih Liu 4 , Chenhua Zhang 4 , Wenbing Xie 3 , Kunqian Yu 2 , Hong Ding 2, 5 , Kehao Zhao 6 , Shijie Chen 2 , Hualiang Jiang 2 , Stephen B Baylin 3 , Cheng Luo 2
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-02-08 , DOI: 10.1021/acschembio.7b00890 Fei Ye 1, 2 , Xiangqian Kong 3 , Hao Zhang 2 , Yan Liu 2 , Zhiyuan Shao 2 , Jia Jin 1 , Yi Cai 3 , Rukang Zhang 2 , Linjuan Li 2 , Yang W Zhang 3 , Yu-Chih Liu 4 , Chenhua Zhang 4 , Wenbing Xie 3 , Kunqian Yu 2 , Hong Ding 2, 5 , Kehao Zhao 6 , Shijie Chen 2 , Hualiang Jiang 2 , Stephen B Baylin 3 , Cheng Luo 2
Affiliation
|
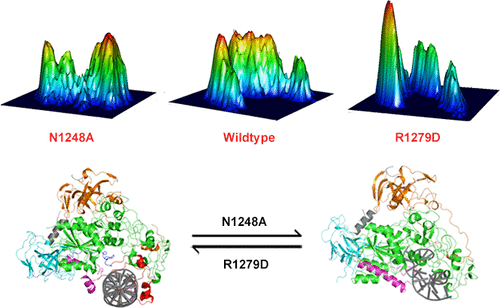
DNA methyltransferase-1 (DNMT1) plays a crucial role in the maintenance of genomic methylation patterns. The crystal structure of DNMT1 was determined in two different states in which the helix that follows the catalytic loop was either kinked (designated helix-kinked) or well folded (designated helix-straight state). Here, we show that the proper structural transition between these two states is required for DNMT1 activity. The mutations of N1248A and R1279D, which did not affect interactions between DNMT1 and substrates or cofactors, allosterically reduced enzymatic activities in vitro by decreasing kcat/ Km for AdoMet. The crystallographic data combined with molecular dynamic (MD) simulations indicated that the N1248A and R1279D mutants bias the catalytic helix to either the kinked or straight conformation. In addition, genetic complementation assays for the two mutants suggested that disturbing the conformational transition reduced DNMT1 activity in cells, which could act additively with existing DNMT inhibitors to decrease DNA methylation. Collectively, our studies provide molecular insights into conformational changes of the catalytic helix, which is essential for DNMT1 catalytic activity, and thus aid in better understanding the relationship between DNMT1 dynamic switching and enzymatic activity.
中文翻译:

生化研究和分子动力学模拟揭示了DNA Methyltransferase-1构象变化的分子基础。
DNA甲基转移酶-1(DNMT1)在维持基因组甲基化模式中起着至关重要的作用。DNMT1的晶体结构是在两种不同的状态下确定的,其中催化环后面的螺旋要么扭结(指定为螺旋扭结),要么折叠得很好(指定为螺旋直状态)。在这里,我们表明DNMT1活动需要在这两个状态之间进行适当的结构转换。N1248A和R1279D的突变不影响DNMT1与底物或辅因子之间的相互作用,但通过降低AdoMet的kcat / Km在体外变构地降低了酶的活性。晶体学数据与分子动力学(MD)模拟相结合表明,N1248A和R1279D突变体使催化螺旋偏向扭结或直构象。此外,这两个突变体的遗传互补分析表明,干扰构象转变会降低细胞中的DNMT1活性,这可能与现有DNMT抑制剂相加作用以减少DNA甲基化。总的来说,我们的研究为DNMT1催化活性必不可少的催化螺旋的构象变化提供了分子见解,因此有助于更好地理解DNMT1动态转换与酶促活性之间的关系。
更新日期:2018-01-30
中文翻译:

生化研究和分子动力学模拟揭示了DNA Methyltransferase-1构象变化的分子基础。
DNA甲基转移酶-1(DNMT1)在维持基因组甲基化模式中起着至关重要的作用。DNMT1的晶体结构是在两种不同的状态下确定的,其中催化环后面的螺旋要么扭结(指定为螺旋扭结),要么折叠得很好(指定为螺旋直状态)。在这里,我们表明DNMT1活动需要在这两个状态之间进行适当的结构转换。N1248A和R1279D的突变不影响DNMT1与底物或辅因子之间的相互作用,但通过降低AdoMet的kcat / Km在体外变构地降低了酶的活性。晶体学数据与分子动力学(MD)模拟相结合表明,N1248A和R1279D突变体使催化螺旋偏向扭结或直构象。此外,这两个突变体的遗传互补分析表明,干扰构象转变会降低细胞中的DNMT1活性,这可能与现有DNMT抑制剂相加作用以减少DNA甲基化。总的来说,我们的研究为DNMT1催化活性必不可少的催化螺旋的构象变化提供了分子见解,因此有助于更好地理解DNMT1动态转换与酶促活性之间的关系。











































 京公网安备 11010802027423号
京公网安备 11010802027423号