当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Accuracy Comparison of Generalized Born Models in the Calculation of Electrostatic Binding Free Energies
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-01-29 00:00:00 , DOI: 10.1021/acs.jctc.7b00886 Saeed Izadi 1 , Robert C. Harris 2 , Marcia O. Fenley 3 , Alexey V. Onufriev
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-01-29 00:00:00 , DOI: 10.1021/acs.jctc.7b00886 Saeed Izadi 1 , Robert C. Harris 2 , Marcia O. Fenley 3 , Alexey V. Onufriev
Affiliation
|
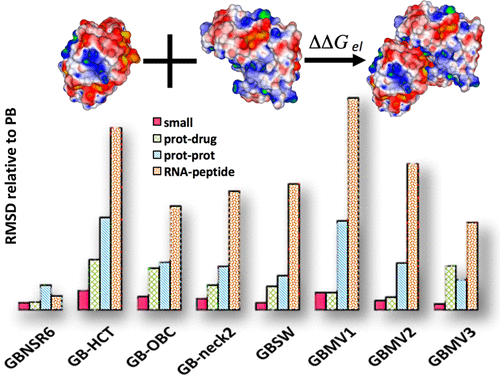
The need for accurate yet efficient representation of the aqueous environment in biomolecular modeling has led to the development of a variety of generalized Born (GB) implicit solvent models. While many studies have focused on the accuracy of available GB models in predicting solvation free energies, a systematic assessment of the quality of these models in binding free energy calculations, crucial for rational drug design, has not been undertaken. Here, we evaluate the accuracies of eight common GB flavors (GB-HCT, GB-OBC, GB-neck2, GBNSR6, GBSW, GBMV1, GBMV2, and GBMV3), available in major molecular dynamics packages, in predicting the electrostatic binding free energies (ΔΔGel) for a diverse set of 60 biomolecular complexes belonging to four main classes: protein–protein, protein-drug, RNA-peptide, and small complexes. The GB flavors are examined in terms of their ability to reproduce the results from the Poisson–Boltzmann (PB) model, commonly used as accuracy reference in this context. We show that the agreement with the PB of ΔΔGel estimates varies widely between different GB models and also across different types of biomolecular complexes, with R2 correlations ranging from 0.3772 to 0.9986. A surface-based “R6” GB model recently implemented in AMBER shows the closest overall agreement with reference PB (R2 = 0.9949, RMSD = 8.75 kcal/mol). The RNA-peptide and protein-drug complex sets appear to be most challenging for all but one model, as indicated by the large deviations from the PB in ΔΔGel. Small neutral complexes present the least challenge for most of the GB models tested. The quantitative demonstration of the strengths and weaknesses of the GB models across the diverse complex types provided here can be used as a guide for practical computations and future development efforts.
中文翻译:

广义Born模型在静电自由结合能计算中的精度比较。
在生物分子建模中需要精确而有效地表示水环境的需求导致了各种广义Born(GB)隐式溶剂模型的开发。尽管许多研究集中在可用GB模型预测溶剂化自由能的准确性上,但尚未进行对结合自由能计算中这些模型的质量的系统评估,这对于合理的药物设计至关重要。在这里,我们评估了主要分子动力学软件包中可用的八种常见GB口味(GB-HCT,GB-OBC,GB-neck2,GBNSR6,GBSW,GBMV1,GBMV2和GBMV3)的准确性,以预测静电结合自由能(ΔΔ ģ EL),涵盖了60种生物分子复合物,分别属于四个主要类别:蛋白质-蛋白质,蛋白质-药物,RNA肽和小型复合物。对GB香精进行了重现Poisson-Boltzmann(PB)模型结果的能力的检验,该模型通常在此情况下用作准确性参考。我们表明,的PB协议ΔΔ摹EL估计变化不同GB型号之间广泛,也跨越不同类型的生物分子复合物,与[R 2间的相关性,从0.3772到0.9986。最近在AMBER中实施的基于表面的“ R6” GB模型显示了与参考PB(R 2= 0.9949,RMSD = 8.75 kcal / mol)。所述RNA-肽和蛋白质-药物复合套似乎是最有挑战性的,但所有一个模型,通过从PB大偏差的所指示的ΔΔ ģ EL。对于大多数测试的GB模型,小型中性配合物带来的挑战最少。此处提供的有关各种复杂类型的GB模型的优缺点的定量演示可以用作实际计算和未来开发工作的指南。
更新日期:2018-01-29
中文翻译:

广义Born模型在静电自由结合能计算中的精度比较。
在生物分子建模中需要精确而有效地表示水环境的需求导致了各种广义Born(GB)隐式溶剂模型的开发。尽管许多研究集中在可用GB模型预测溶剂化自由能的准确性上,但尚未进行对结合自由能计算中这些模型的质量的系统评估,这对于合理的药物设计至关重要。在这里,我们评估了主要分子动力学软件包中可用的八种常见GB口味(GB-HCT,GB-OBC,GB-neck2,GBNSR6,GBSW,GBMV1,GBMV2和GBMV3)的准确性,以预测静电结合自由能(ΔΔ ģ EL),涵盖了60种生物分子复合物,分别属于四个主要类别:蛋白质-蛋白质,蛋白质-药物,RNA肽和小型复合物。对GB香精进行了重现Poisson-Boltzmann(PB)模型结果的能力的检验,该模型通常在此情况下用作准确性参考。我们表明,的PB协议ΔΔ摹EL估计变化不同GB型号之间广泛,也跨越不同类型的生物分子复合物,与[R 2间的相关性,从0.3772到0.9986。最近在AMBER中实施的基于表面的“ R6” GB模型显示了与参考PB(R 2= 0.9949,RMSD = 8.75 kcal / mol)。所述RNA-肽和蛋白质-药物复合套似乎是最有挑战性的,但所有一个模型,通过从PB大偏差的所指示的ΔΔ ģ EL。对于大多数测试的GB模型,小型中性配合物带来的挑战最少。此处提供的有关各种复杂类型的GB模型的优缺点的定量演示可以用作实际计算和未来开发工作的指南。











































 京公网安备 11010802027423号
京公网安备 11010802027423号