当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Analyzing and Tuning Ribozyme Activity by Deep Sequencing To Modulate Gene Expression Level in Mammalian Cells
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-01-24 00:00:00 , DOI: 10.1021/acssynbio.7b00367 Shungo Kobori 1 , Yohei Yokobayashi 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-01-24 00:00:00 , DOI: 10.1021/acssynbio.7b00367 Shungo Kobori 1 , Yohei Yokobayashi 1
Affiliation
|
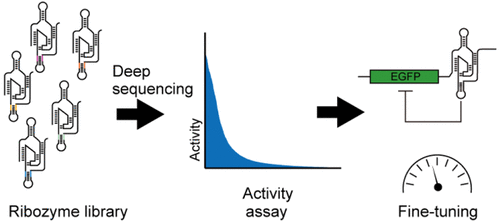
Self-cleaving ribozymes, in combination with aptamers and various classes of RNAs, have been heavily engineered to create RNA devices to control gene expression. Although understanding of sequence–function relationships of ribozymes is critical for such efforts, our current knowledge of self-cleaving ribozymes is mostly limited to the results from small scale mutational studies performed under different conditions, or qualitative results of mutate-and-select experiments that may contain experimental biases. Here, we applied our strategy based on deep sequencing to comprehensively assay a large number of mutants to systematically examine the effect of the P4 stem sequence on the activity of an HDV-like ribozyme. We discovered that the ribozyme activity is highly sensitive to the sequence and the apparent stability of the varied positions. Furthermore, we demonstrated that the collection of the ribozyme variants with different activities can be used as a convenient device to fine-tune the level of gene expression in mammalian cells.
中文翻译:

通过深度测序分析和调节核酶活性,以调节哺乳动物细胞中的基因表达水平
自裂解核酶,与适体和各种类型的RNA结合,已被大量工程化,以创建可控制基因表达的RNA装置。尽管了解核酶的序列-功能关系对于此类工作至关重要,但我们目前对自切割核酶的了解主要限于在不同条件下进行的小规模突变研究的结果,或定性和选择性实验的结果。可能包含实验偏差。在这里,我们应用了基于深度测序的策略来全面分析大量突变体,以系统地检查P4茎序列对HDV样核酶活性的影响。我们发现核酶活性对序列和不同位置的表观稳定性高度敏感。
更新日期:2018-01-24
中文翻译:

通过深度测序分析和调节核酶活性,以调节哺乳动物细胞中的基因表达水平
自裂解核酶,与适体和各种类型的RNA结合,已被大量工程化,以创建可控制基因表达的RNA装置。尽管了解核酶的序列-功能关系对于此类工作至关重要,但我们目前对自切割核酶的了解主要限于在不同条件下进行的小规模突变研究的结果,或定性和选择性实验的结果。可能包含实验偏差。在这里,我们应用了基于深度测序的策略来全面分析大量突变体,以系统地检查P4茎序列对HDV样核酶活性的影响。我们发现核酶活性对序列和不同位置的表观稳定性高度敏感。









































 京公网安备 11010802027423号
京公网安备 11010802027423号