Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Restriction Enzymes as a Target for DNA-Based Sensing and Structural Rearrangement
ACS Omega ( IF 3.7 ) Pub Date : 2018-01-17 00:00:00 , DOI: 10.1021/acsomega.7b01333 Susan Buckhout-White 1 , Chanel Person 1 , Igor L Medintz 1 , Ellen R Goldman 1
ACS Omega ( IF 3.7 ) Pub Date : 2018-01-17 00:00:00 , DOI: 10.1021/acsomega.7b01333 Susan Buckhout-White 1 , Chanel Person 1 , Igor L Medintz 1 , Ellen R Goldman 1
Affiliation
|
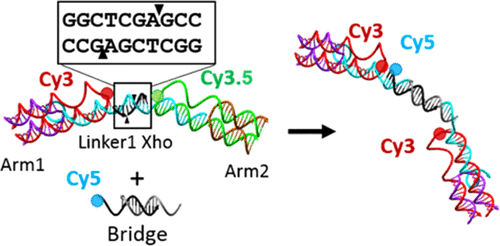
DNA nanostructures have been shown viable for the creation of complex logic-enabled sensing motifs. To date, most of these types of devices have been limited to the interaction with strictly DNA-type inputs. Restriction endonuclease represents a class of enzyme with endogenous specificity to DNA, and we hypothesize that these can be integrated with a DNA structure for use as inputs to trigger structural transformation and structural rearrangement. In this work, we reconfigured a three-arm DNA switch, which utilizes a cyclic Förster resonance energy transfer interaction between three dyes to produce complex output for the detection of three separate input regions to respond to restriction endonucleases, and investigated the efficacy of the enzyme targets. We demonstrate the ability to use three enzymes in one switch with no nonspecific interaction between cleavage sites. Further, we show that the enzymatic digestion can be harnessed to expose an active toehold into the DNA structure, allowing for single-pot addition of a small oligo in solution.
中文翻译:

限制性内切酶作为基于 DNA 的传感和结构重排的靶标
DNA 纳米结构已被证明可用于创建复杂的逻辑传感图案。迄今为止,大多数此类设备仅限于与严格的 DNA 类型输入进行交互。限制性内切酶代表一类对 DNA 具有内源特异性的酶,我们假设它们可以与 DNA 结构整合,用作触发结构转化和结构重排的输入。在这项工作中,我们重新配置了一个三臂 DNA 开关,它利用三种染料之间的循环福斯特共振能量转移相互作用来产生复杂的输出,用于检测三个独立的输入区域以响应限制性内切核酸酶,并研究了该酶的功效目标。我们证明了在一个开关中使用三种酶的能力,并且切割位点之间没有非特异性相互作用。此外,我们还表明,可以利用酶消化将活性立足点暴露到 DNA 结构中,从而允许在溶液中单锅添加小型寡核苷酸。
更新日期:2018-01-17
中文翻译:

限制性内切酶作为基于 DNA 的传感和结构重排的靶标
DNA 纳米结构已被证明可用于创建复杂的逻辑传感图案。迄今为止,大多数此类设备仅限于与严格的 DNA 类型输入进行交互。限制性内切酶代表一类对 DNA 具有内源特异性的酶,我们假设它们可以与 DNA 结构整合,用作触发结构转化和结构重排的输入。在这项工作中,我们重新配置了一个三臂 DNA 开关,它利用三种染料之间的循环福斯特共振能量转移相互作用来产生复杂的输出,用于检测三个独立的输入区域以响应限制性内切核酸酶,并研究了该酶的功效目标。我们证明了在一个开关中使用三种酶的能力,并且切割位点之间没有非特异性相互作用。此外,我们还表明,可以利用酶消化将活性立足点暴露到 DNA 结构中,从而允许在溶液中单锅添加小型寡核苷酸。











































 京公网安备 11010802027423号
京公网安备 11010802027423号