当前位置:
X-MOL 学术
›
J. Nat. Prod.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification and Proposed Relative and Absolute Configurations of Niphimycins C–E from the Marine-Derived Streptomyces sp. IMB7-145 by Genomic Analysis
Journal of Natural Products ( IF 5.1 ) Pub Date : 2018-01-08 00:00:00 , DOI: 10.1021/acs.jnatprod.7b00859 Yuanyuan Hu 1 , Mian Wang 1 , Chunyan Wu 1, 2 , Yi Tan 1 , Jiao Li 1 , Xiaomeng Hao 1 , Yanbo Duan 1 , Yan Guan 1 , Xiaoya Shang 2 , Yiguang Wang 1 , Chunling Xiao 1 , Maoluo Gan 1
Journal of Natural Products ( IF 5.1 ) Pub Date : 2018-01-08 00:00:00 , DOI: 10.1021/acs.jnatprod.7b00859 Yuanyuan Hu 1 , Mian Wang 1 , Chunyan Wu 1, 2 , Yi Tan 1 , Jiao Li 1 , Xiaomeng Hao 1 , Yanbo Duan 1 , Yan Guan 1 , Xiaoya Shang 2 , Yiguang Wang 1 , Chunling Xiao 1 , Maoluo Gan 1
Affiliation
|
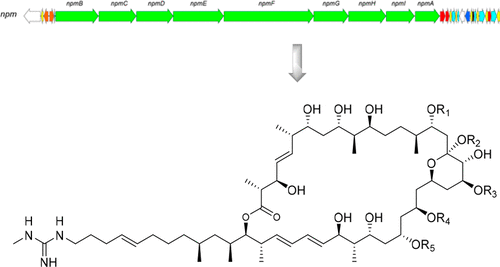
Analysis of the whole genome sequence of Streptomyces sp. IMB7-145 revealed the presence of seven type I polyketide synthase biosynthetic gene clusters, one of which was highly homologous to the biosynthetic gene cluster of azalomycin F. Detailed bioinformatic analysis of the modular organization of the PKS gene suggested that this gene is responsible for niphimycin biosynthesis. Guided by genomic analysis, a large-scale cultivation ultimately led to the discovery and characterization of four new niphimycin congeners, namely, niphimycins C–E (1–3) and 17-O-methylniphimycin (4). The configurations of most stereocenters of niphimycins have not been determined to date. In the present study, the relative configurations were elucidated by spectroscopic analysis, including J-based analysis and the CNMR database method. Further, the full absolute configurations of niphimycins were completely proposed for the first time based on biosynthetic gene cluster analysis of the ketoreductase and enoylreductase domains for hydroxy- and methyl-bearing stereocenters. Compounds 1, 3, 4, and niphimycin Iα (5) showed antimicrobial activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci (MIC: 8–64 μg/mL), as well as cytotoxicity against the human HeLa cancer cell line (IC50: 3.0–9.0 μM). In addition, compounds 1 and 5 displayed significant activity against several Mycobacterium tuberculosis clinical isolates (MIC: 4–32 μg/mL).
中文翻译:

从海洋衍生链霉菌属菌种鉴定和拟议的相对分子构型和相对构型的Niphimys C–E 。IMB7-145通过基因组分析
链霉菌的全基因组序列分析。IMB7-145揭示了七个I型聚酮化合物合酶生物合成基因簇的存在,其中之一与阿扎霉素F的生物合成基因簇高度同源。对PKS基因模块结构的详细生物信息学分析表明,该基因负责尼福霉素生物合成。通过基因组分析的指导下,一个大规模培养最终导致了四个新niphimycin同源的发现和表征,即,niphimycins C-E(1 - 3)和17- ø -methylniphimycin(4)。迄今为止,尚未确定大多数霉素的立体中心的构型。在本研究中,通过光谱分析阐明了相对构型,包括基于J的分析和CNMR数据库方法。此外,基于对羟基和甲基的立体中心的酮还原酶和烯酰还原酶结构域的生物合成基因簇分析,首次首次提出了尼福霉素的完全绝对构型。化合物1,3,4,和niphimycinIα(5)显示出抗微生物活性对耐甲氧西林金黄色葡萄球菌耐万古霉素的肠球菌(MIC:8–64μg/ mL),以及对人类HeLa癌细胞系的细胞毒性(IC 50:3.0–9.0μM)。此外,化合物1和5对几种结核分枝杆菌临床分离株(MIC:4–32μg/ mL)显示出显着活性。
更新日期:2018-01-08
中文翻译:

从海洋衍生链霉菌属菌种鉴定和拟议的相对分子构型和相对构型的Niphimys C–E 。IMB7-145通过基因组分析
链霉菌的全基因组序列分析。IMB7-145揭示了七个I型聚酮化合物合酶生物合成基因簇的存在,其中之一与阿扎霉素F的生物合成基因簇高度同源。对PKS基因模块结构的详细生物信息学分析表明,该基因负责尼福霉素生物合成。通过基因组分析的指导下,一个大规模培养最终导致了四个新niphimycin同源的发现和表征,即,niphimycins C-E(1 - 3)和17- ø -methylniphimycin(4)。迄今为止,尚未确定大多数霉素的立体中心的构型。在本研究中,通过光谱分析阐明了相对构型,包括基于J的分析和CNMR数据库方法。此外,基于对羟基和甲基的立体中心的酮还原酶和烯酰还原酶结构域的生物合成基因簇分析,首次首次提出了尼福霉素的完全绝对构型。化合物1,3,4,和niphimycinIα(5)显示出抗微生物活性对耐甲氧西林金黄色葡萄球菌耐万古霉素的肠球菌(MIC:8–64μg/ mL),以及对人类HeLa癌细胞系的细胞毒性(IC 50:3.0–9.0μM)。此外,化合物1和5对几种结核分枝杆菌临床分离株(MIC:4–32μg/ mL)显示出显着活性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号