Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Concentrating and labeling genomic DNA in a nanofluidic array†
Nanoscale ( IF 5.8 ) Pub Date : 2018-01-04 00:00:00 , DOI: 10.1039/c7nr06016e Rodolphe Marie 1, 2, 3, 4 , Jonas N. Pedersen 1, 2, 3, 4 , Kalim U. Mir 5, 6, 7, 8 , Brian Bilenberg 3, 4, 9 , Anders Kristensen 1, 2, 3, 4
Nanoscale ( IF 5.8 ) Pub Date : 2018-01-04 00:00:00 , DOI: 10.1039/c7nr06016e Rodolphe Marie 1, 2, 3, 4 , Jonas N. Pedersen 1, 2, 3, 4 , Kalim U. Mir 5, 6, 7, 8 , Brian Bilenberg 3, 4, 9 , Anders Kristensen 1, 2, 3, 4
Affiliation
|
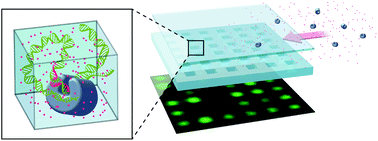
Nucleotide incorporation by DNA polymerase forms the basis of DNA sequencing-by-synthesis. In current platforms, either the single-stranded DNA or the enzyme is immobilized on a solid surface to locate the incorporation of individual nucleotides in space and/or time. Solid-phase reactions may, however, hinder the polymerase activity. We demonstrate a device and a protocol for the enzymatic labeling of genomic DNA arranged in a dense array of single molecules without attaching the enzyme or the DNA to a surface. DNA molecules accumulate in a dense array of pits embedded within a nanoslit due to entropic trapping. We then perform ϕ29 polymerase extension from single-strand nicks created on the trapped molecules to incorporate fluorescent nucleotides into the DNA. The array of entropic traps can be loaded with λ-DNA molecules to more than 90% of capacity at a flow rate of 10 pL min−1. The final concentration can reach up to 100 μg mL−1, and the DNA is eluted from the array by increasing the flow rate. The device may be an important preparative module for carrying out enzymatic processing on DNA extracted from single-cells in a microfluidic chip.
中文翻译:

在纳米流体阵列中浓缩和标记基因组DNA †
DNA聚合酶掺入核苷酸构成了DNA合成测序的基础。在当前平台中,单链DNA或酶被固定在固体表面上,以定位单个核苷酸在空间和/或时间上的掺入。但是,固相反应可能会阻碍聚合酶的活性。我们展示了一种酶和标记的基因组DNA排列在单个分子的密集阵列中而不附加酶或DNA到表面的设备和协议。由于熵陷阱,DNA分子堆积在纳米缝隙中密集的凹坑阵列中。然后我们执行ϕ从被困分子上产生的单链缺口开始29种聚合酶延伸,以将荧光核苷酸整合到DNA中。熵阱的阵列可以以10 pL min -1的流速装载λ-DNA分子至90%以上的容量。最终浓度可以达到100μgmL -1,并且可以通过增加流速从阵列中洗脱DNA。该设备可能是重要的制备模块,用于对微流控芯片中从单细胞提取的DNA进行酶促处理。
更新日期:2018-01-04
中文翻译:

在纳米流体阵列中浓缩和标记基因组DNA †
DNA聚合酶掺入核苷酸构成了DNA合成测序的基础。在当前平台中,单链DNA或酶被固定在固体表面上,以定位单个核苷酸在空间和/或时间上的掺入。但是,固相反应可能会阻碍聚合酶的活性。我们展示了一种酶和标记的基因组DNA排列在单个分子的密集阵列中而不附加酶或DNA到表面的设备和协议。由于熵陷阱,DNA分子堆积在纳米缝隙中密集的凹坑阵列中。然后我们执行ϕ从被困分子上产生的单链缺口开始29种聚合酶延伸,以将荧光核苷酸整合到DNA中。熵阱的阵列可以以10 pL min -1的流速装载λ-DNA分子至90%以上的容量。最终浓度可以达到100μgmL -1,并且可以通过增加流速从阵列中洗脱DNA。该设备可能是重要的制备模块,用于对微流控芯片中从单细胞提取的DNA进行酶促处理。











































 京公网安备 11010802027423号
京公网安备 11010802027423号