当前位置:
X-MOL 学术
›
Chem. Rev.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview.
Chemical Reviews ( IF 51.4 ) Pub Date : 2018-01-03 , DOI: 10.1021/acs.chemrev.7b00427 Jiří Šponer 1 , Giovanni Bussi 2 , Miroslav Krepl 1, 3 , Pavel Banáš 3 , Sandro Bottaro 4 , Richard A Cunha 2 , Alejandro Gil-Ley 2 , Giovanni Pinamonti 2 , Simón Poblete 2 , Petr Jurečka 3 , Nils G Walter 5 , Michal Otyepka 3
Chemical Reviews ( IF 51.4 ) Pub Date : 2018-01-03 , DOI: 10.1021/acs.chemrev.7b00427 Jiří Šponer 1 , Giovanni Bussi 2 , Miroslav Krepl 1, 3 , Pavel Banáš 3 , Sandro Bottaro 4 , Richard A Cunha 2 , Alejandro Gil-Ley 2 , Giovanni Pinamonti 2 , Simón Poblete 2 , Petr Jurečka 3 , Nils G Walter 5 , Michal Otyepka 3
Affiliation
|
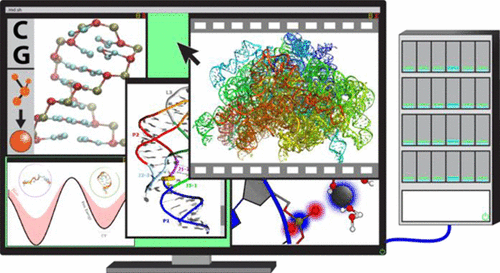
With both catalytic and genetic functions, ribonucleic acid (RNA) is perhaps the most pluripotent chemical species in molecular biology, and its functions are intimately linked to its structure and dynamics. Computer simulations, and in particular atomistic molecular dynamics (MD), allow structural dynamics of biomolecular systems to be investigated with unprecedented temporal and spatial resolution. We here provide a comprehensive overview of the fast-developing field of MD simulations of RNA molecules. We begin with an in-depth, evaluatory coverage of the most fundamental methodological challenges that set the basis for the future development of the field, in particular, the current developments and inherent physical limitations of the atomistic force fields and the recent advances in a broad spectrum of enhanced sampling methods. We also survey the closely related field of coarse-grained modeling of RNA systems. After dealing with the methodological aspects, we provide an exhaustive overview of the available RNA simulation literature, ranging from studies of the smallest RNA oligonucleotides to investigations of the entire ribosome. Our review encompasses tetranucleotides, tetraloops, a number of small RNA motifs, A-helix RNA, kissing-loop complexes, the TAR RNA element, the decoding center and other important regions of the ribosome, as well as assorted others systems. Extended sections are devoted to RNA-ion interactions, ribozymes, riboswitches, and protein/RNA complexes. Our overview is written for as broad of an audience as possible, aiming to provide a much-needed interdisciplinary bridge between computation and experiment, together with a perspective on the future of the field.
中文翻译:

分子模拟捕获的 RNA 结构动力学:综合概述。
核糖核酸(RNA)兼具催化和遗传功能,可能是分子生物学中最具多能性的化学物质,其功能与其结构和动力学密切相关。计算机模拟,特别是原子分子动力学(MD),允许以前所未有的时间和空间分辨率研究生物分子系统的结构动力学。我们在此全面概述快速发展的 RNA 分子 MD 模拟领域。我们首先对最基本的方法论挑战进行深入的评估,这些挑战为该领域的未来发展奠定了基础,特别是原子力场的当前发展和固有的物理局限性以及广泛领域的最新进展。一系列增强采样方法。我们还调查了密切相关的 RNA 系统粗粒度建模领域。在讨论了方法论方面之后,我们对现有的 RNA 模拟文献进行了详尽的概述,范围从最小的 RNA 寡核苷酸的研究到整个核糖体的研究。我们的综述涵盖四核苷酸、四环、许多小 RNA 基序、A 螺旋 RNA、接吻环复合物、TAR RNA 元件、解码中心和核糖体的其他重要区域,以及各种其他系统。扩展部分专门讨论 RNA-离子相互作用、核酶、核糖开关和蛋白质/RNA 复合物。我们的概述是为尽可能广泛的受众编写的,旨在提供计算和实验之间急需的跨学科桥梁,以及对该领域未来的展望。
更新日期:2018-01-03
中文翻译:

分子模拟捕获的 RNA 结构动力学:综合概述。
核糖核酸(RNA)兼具催化和遗传功能,可能是分子生物学中最具多能性的化学物质,其功能与其结构和动力学密切相关。计算机模拟,特别是原子分子动力学(MD),允许以前所未有的时间和空间分辨率研究生物分子系统的结构动力学。我们在此全面概述快速发展的 RNA 分子 MD 模拟领域。我们首先对最基本的方法论挑战进行深入的评估,这些挑战为该领域的未来发展奠定了基础,特别是原子力场的当前发展和固有的物理局限性以及广泛领域的最新进展。一系列增强采样方法。我们还调查了密切相关的 RNA 系统粗粒度建模领域。在讨论了方法论方面之后,我们对现有的 RNA 模拟文献进行了详尽的概述,范围从最小的 RNA 寡核苷酸的研究到整个核糖体的研究。我们的综述涵盖四核苷酸、四环、许多小 RNA 基序、A 螺旋 RNA、接吻环复合物、TAR RNA 元件、解码中心和核糖体的其他重要区域,以及各种其他系统。扩展部分专门讨论 RNA-离子相互作用、核酶、核糖开关和蛋白质/RNA 复合物。我们的概述是为尽可能广泛的受众编写的,旨在提供计算和实验之间急需的跨学科桥梁,以及对该领域未来的展望。











































 京公网安备 11010802027423号
京公网安备 11010802027423号