Bioorganic & Medicinal Chemistry ( IF 3.3 ) Pub Date : 2017-12-26 , DOI: 10.1016/j.bmc.2017.12.040 Xue-Song Wang , Qing-Chuan Zheng
|
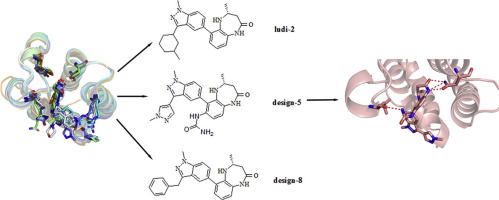
The CBP (CREB (cAMP responsive element binding protein) binding protein) bromodomain (BRD) could recognize and bind with acetyl K382 of human tumor suppressor protein p53 which the mutation of encoding gene might cause human cancers. CBP-BRD serves as a promising drug target for several disease pathways and a series of effective drug have been discovered. In this study, molecular dynamics (MD) simulations and molecular mechanics generalized born surface area (MM-GB/SA) approaches were performed to investigate the different binding modes between five inhibitors with CBP-BRD. Based on the energy and conformation analyses, a potent core fragment is chosen to act as the starting point for new inhibitor design by means of LUDI and rational drug design approaches. Then, T.E.S.T and molinspirition were applied to evaluate oral bioavailability and drug promiscuity of the new molecules. These results shed light on the idea for further inhibitor design.
中文翻译:

不同抑制剂结构特征及与CBP溴结构域结合方式差异的理论研究
CBP(CREB(cAMP响应元件结合蛋白)结合蛋白)溴结构域(BRD)可以识别并与人类抑癌蛋白p53的乙酰K382结合,编码基因的突变可能导致人类癌症。CBP-BRD作为多种疾病途径的有希望的药物靶标,已经发现了一系列有效的药物。在这项研究中,进行了分子动力学(MD)模拟和分子力学广义生表面积(MM-GB / SA)方法,以研究五种抑制剂与CBP-BRD的不同结合方式。基于能量和构象分析,通过LUDI和合理的药物设计方法,选择有效的核心片段作为新抑制剂设计的起点。然后,TES T和molinspirition用于评估新分子的口服生物利用度和药物滥用。这些结果为进一步的抑制剂设计提供了思路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号