当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mol2vec: Unsupervised Machine Learning Approach with Chemical Intuition
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-10 00:00:00 , DOI: 10.1021/acs.jcim.7b00616 Sabrina Jaeger 1 , Simone Fulle 1 , Samo Turk 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-10 00:00:00 , DOI: 10.1021/acs.jcim.7b00616 Sabrina Jaeger 1 , Simone Fulle 1 , Samo Turk 1
Affiliation
|
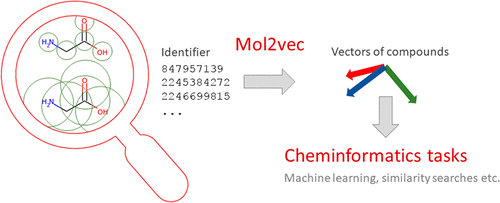
Inspired by natural language processing techniques, we here introduce Mol2vec, which is an unsupervised machine learning approach to learn vector representations of molecular substructures. Like the Word2vec models, where vectors of closely related words are in close proximity in the vector space, Mol2vec learns vector representations of molecular substructures that point in similar directions for chemically related substructures. Compounds can finally be encoded as vectors by summing the vectors of the individual substructures and, for instance, be fed into supervised machine learning approaches to predict compound properties. The underlying substructure vector embeddings are obtained by training an unsupervised machine learning approach on a so-called corpus of compounds that consists of all available chemical matter. The resulting Mol2vec model is pretrained once, yields dense vector representations, and overcomes drawbacks of common compound feature representations such as sparseness and bit collisions. The prediction capabilities are demonstrated on several compound property and bioactivity data sets and compared with results obtained for Morgan fingerprints as a reference compound representation. Mol2vec can be easily combined with ProtVec, which employs the same Word2vec concept on protein sequences, resulting in a proteochemometric approach that is alignment-independent and thus can also be easily used for proteins with low sequence similarities.
中文翻译:

Mol2vec:具有化学直觉的无监督机器学习方法
受自然语言处理技术的启发,我们在这里介绍Mol2vec,这是一种无监督的机器学习方法,用于学习分子亚结构的向量表示。就像Word2vec模型一样,密切相关的单词的向量在向量空间中非常接近,Mol2vec可以学习指向化学相关子结构的相似方向的分子子结构的向量表示。通过将各个子结构的向量求和,最终可以将化合物编码为向量,例如,可以将其馈入有监督的机器学习方法中以预测化合物的性质。底层的子结构矢量嵌入是通过在所谓的包含所有可用化学物质的化合物主体上训练无监督机器学习方法而获得的。生成的Mol2vec模型经过一次预训练,产生密集的矢量表示,并克服了常见复合特征表示的缺点,例如稀疏性和位冲突。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与作为参考化合物表示形式的Morgan指纹图谱的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与以Morgan指纹图谱作为参考化合物表示形式获得的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与以Morgan指纹图谱作为参考化合物表示形式获得的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。
更新日期:2018-01-10
中文翻译:

Mol2vec:具有化学直觉的无监督机器学习方法
受自然语言处理技术的启发,我们在这里介绍Mol2vec,这是一种无监督的机器学习方法,用于学习分子亚结构的向量表示。就像Word2vec模型一样,密切相关的单词的向量在向量空间中非常接近,Mol2vec可以学习指向化学相关子结构的相似方向的分子子结构的向量表示。通过将各个子结构的向量求和,最终可以将化合物编码为向量,例如,可以将其馈入有监督的机器学习方法中以预测化合物的性质。底层的子结构矢量嵌入是通过在所谓的包含所有可用化学物质的化合物主体上训练无监督机器学习方法而获得的。生成的Mol2vec模型经过一次预训练,产生密集的矢量表示,并克服了常见复合特征表示的缺点,例如稀疏性和位冲突。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与作为参考化合物表示形式的Morgan指纹图谱的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与以Morgan指纹图谱作为参考化合物表示形式获得的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。在几个化合物特性和生物活性数据集上证明了预测能力,并将其与以Morgan指纹图谱作为参考化合物表示形式获得的结果进行了比较。Mol2vec可以轻松地与ProtVec结合使用,后者对蛋白质序列采用相同的Word2vec概念,从而产生了一种蛋白质化学计量学方法,该方法不依赖于比对,因此也可以轻松用于序列相似性低的蛋白质。










































 京公网安备 11010802027423号
京公网安备 11010802027423号