当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Complexity in pH-Dependent Ribozyme Kinetics: Dark pKa Shifts and Wavy Rate–pH Profiles
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acs.biochem.7b00784 Erica A. Frankel 1, 2 , Philip C. Bevilacqua 1, 2, 3
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-22 00:00:00 , DOI: 10.1021/acs.biochem.7b00784 Erica A. Frankel 1, 2 , Philip C. Bevilacqua 1, 2, 3
Affiliation
|
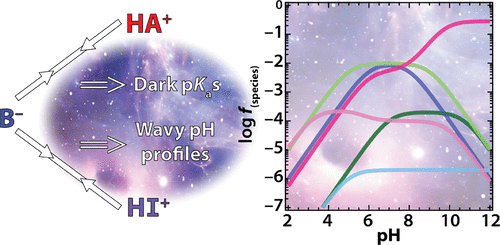
Charged bases occur in RNA enzymes, or ribozymes, where they play key roles in catalysis. Cationic bases donate protons and perform electrostatic catalysis, while anionic bases accept protons. We previously published simulations of rate–pH profiles for ribozymes in terms of species plots for the general acid and general base that have been useful for understanding how ribozymes respond to pH. In that study, we did not consider interaction between the general acid and general base or interaction with other species on the RNA. Since that report, diverse small ribozyme classes have been discovered, many of which have charged nucleobases or metal ions in the active site that can either directly interact and participate in catalysis or indirectly interact as “influencers”. Herein, we simulate experimental rate–pH profiles in terms of species plots in which reverse protonated charged nucleobases interact. These analyses uncover two surprising features of pH-dependent enzyme kinetics. (1) Cooperativity between the general acid and general base enhances population of the functional forms of a ribozyme and manifests itself as hidden or “dark” pKa shifts, real pKa shifts that accelerate the reaction but are not readily observed by standard experimental approaches, and (2) influencers favorably shift the pKas of proton-transferring nucleobases and manifest themselves as “wavy” rate–pH profiles. We identify parallels with the protein enzyme literature, including reverse protonation and wavelike behavior, while pointing out that RNA is more prone to reverse protonation. The complexities uncovered, which arise from simple pairwise interactions, should aid deconvolution of complex rate–pH profiles for RNA and protein enzymes and suggest veiled catalytic devices for promoting catalysis that can be tested by experiment and calculation.
中文翻译:

pH依赖的核酶动力学的复杂性:暗的p K a位移和波浪速率–pH谱
带电荷的碱基存在于RNA酶或核酶中,它们在催化中起关键作用。阳离子碱提供质子并进行静电催化,而阴离子碱接受质子。我们以前曾发表过关于核酶的速率-pH曲线的模拟图,该图以一般酸和一般碱的物种图表示,对于理解核酶如何响应pH值非常有用。在该研究中,我们没有考虑通用酸与通用碱之间的相互作用或与RNA上其他物种的相互作用。自从该报告以来,已经发现了各种各样的小核酶,其中许多在活性位点带了核碱基或金属离子,它们可以直接相互作用并参与催化作用,也可以作为“影响者”间接相互作用。在此处,我们用反向质子化带电核碱基相互作用的物种图来模拟实验速率-pH分布图。这些分析揭示了pH依赖性酶动力学的两个令人惊讶的特征。(1)通用酸和通用碱之间的协同作用增强了核酶功能形式的填充,并表现为隐藏或“暗”的pK a位移,实际的p K a位移会加速反应,但无法通过标准实验方法轻易观察到,并且(2)影响者有利地将p K a s转移到质子转移核碱基上,并表现为“波浪”速率-pH个人资料。我们确定了与蛋白质酶文献的相似之处,包括反向质子化和波状行为,同时指出RNA更倾向于反向质子化。由简单的成对相互作用引起的发现的复杂性,应有助于对RNA和蛋白质酶的复杂速率-pH分布图进行反卷积,并建议采用可通过实验和计算进行测试的带阴影的催化装置,以促进催化作用。
更新日期:2017-12-22
中文翻译:

pH依赖的核酶动力学的复杂性:暗的p K a位移和波浪速率–pH谱
带电荷的碱基存在于RNA酶或核酶中,它们在催化中起关键作用。阳离子碱提供质子并进行静电催化,而阴离子碱接受质子。我们以前曾发表过关于核酶的速率-pH曲线的模拟图,该图以一般酸和一般碱的物种图表示,对于理解核酶如何响应pH值非常有用。在该研究中,我们没有考虑通用酸与通用碱之间的相互作用或与RNA上其他物种的相互作用。自从该报告以来,已经发现了各种各样的小核酶,其中许多在活性位点带了核碱基或金属离子,它们可以直接相互作用并参与催化作用,也可以作为“影响者”间接相互作用。在此处,我们用反向质子化带电核碱基相互作用的物种图来模拟实验速率-pH分布图。这些分析揭示了pH依赖性酶动力学的两个令人惊讶的特征。(1)通用酸和通用碱之间的协同作用增强了核酶功能形式的填充,并表现为隐藏或“暗”的pK a位移,实际的p K a位移会加速反应,但无法通过标准实验方法轻易观察到,并且(2)影响者有利地将p K a s转移到质子转移核碱基上,并表现为“波浪”速率-pH个人资料。我们确定了与蛋白质酶文献的相似之处,包括反向质子化和波状行为,同时指出RNA更倾向于反向质子化。由简单的成对相互作用引起的发现的复杂性,应有助于对RNA和蛋白质酶的复杂速率-pH分布图进行反卷积,并建议采用可通过实验和计算进行测试的带阴影的催化装置,以促进催化作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号