当前位置:
X-MOL 学术
›
ACS Cent. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comparative Nucleotide-Dependent Interactome Analysis Reveals Shared and Differential Properties of KRas4a and KRas4b
ACS Central Science ( IF 18.2 ) Pub Date : 2017-12-20 00:00:00 , DOI: 10.1021/acscentsci.7b00440 Xiaoyu Zhang 1 , Ji Cao 1 , Seth P. Miller 1 , Hui Jing 1 , Hening Lin 1, 2
ACS Central Science ( IF 18.2 ) Pub Date : 2017-12-20 00:00:00 , DOI: 10.1021/acscentsci.7b00440 Xiaoyu Zhang 1 , Ji Cao 1 , Seth P. Miller 1 , Hui Jing 1 , Hening Lin 1, 2
Affiliation
|
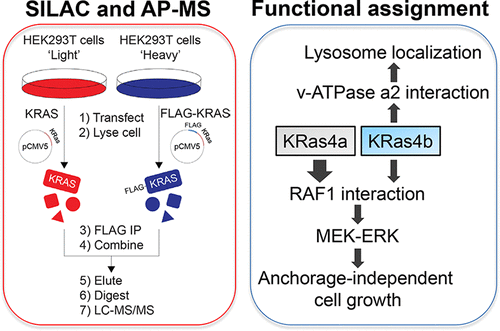
The KRAS gene encodes two isoforms, KRas4a and KRas4b. Differences in the signaling functions of the two KRas proteins are poorly understood. Here we report the comparative and nucleotide-dependent interactomes of KRas4a and KRas4b. Many previously unknown interacting proteins were identified, with some interacting with both isoforms while others prefer only one. For example, v-ATPase a2 and eIF2Bδ interact with only KRas4b. Consistent with the v-ATPase interaction, KRas4b has a significant lysosomal localization. Comparing WT and constitutively active G12D mutant KRas, we examined differences in the effector proteins of the KRas4a and KRas4b. Interestingly, KRas4a binds RAF1 stronger than KRas4b. Correspondingly, KRas4a can better promote ERK phosphorylation and anchorage-independent growth than KRas4b. The interactome data represent a useful resource to understand the differences between KRas4a and KRas4b and to discover new function or regulation for them. A similar proteomic approach would be useful for studying numerous other small GTPases.
中文翻译:

比较核苷酸依赖性的交互作用分析揭示了KRas4a和KRas4b的共享和差异属性。
KRAS基因编码两个同工型,KRas4a和KRas4b。两种KRas蛋白信号传导功能的差异了解甚少。在这里,我们报告比较和核苷酸依赖性的KRas4a和KRas4b的相互作用。已鉴定出许多以前未知的相互作用蛋白,其中一些与两种同工型相互作用,而另一些仅喜欢一种。例如,v-ATPase a2和eIF2Bδ仅与KRas4b相互作用。与v-ATPase相互作用一致,KRas4b具有明显的溶酶体定位。比较WT和组成型活性G12D突变体KRas,我们检查了KRas4a和KRas4b效应蛋白的差异。有趣的是,KRas4a绑定的RAF1比KRas4b绑定的强。相应地,与KRas4b相比,KRas4a可以更好地促进ERK磷酸化和独立于锚定的生长。交互组数据代表了解KRas4a和KRas4b之间的差异并发现它们的新功能或调控的有用资源。相似的蛋白质组学方法对于研究许多其他小GTP酶将很有用。
更新日期:2017-12-20
中文翻译:

比较核苷酸依赖性的交互作用分析揭示了KRas4a和KRas4b的共享和差异属性。
KRAS基因编码两个同工型,KRas4a和KRas4b。两种KRas蛋白信号传导功能的差异了解甚少。在这里,我们报告比较和核苷酸依赖性的KRas4a和KRas4b的相互作用。已鉴定出许多以前未知的相互作用蛋白,其中一些与两种同工型相互作用,而另一些仅喜欢一种。例如,v-ATPase a2和eIF2Bδ仅与KRas4b相互作用。与v-ATPase相互作用一致,KRas4b具有明显的溶酶体定位。比较WT和组成型活性G12D突变体KRas,我们检查了KRas4a和KRas4b效应蛋白的差异。有趣的是,KRas4a绑定的RAF1比KRas4b绑定的强。相应地,与KRas4b相比,KRas4a可以更好地促进ERK磷酸化和独立于锚定的生长。交互组数据代表了解KRas4a和KRas4b之间的差异并发现它们的新功能或调控的有用资源。相似的蛋白质组学方法对于研究许多其他小GTP酶将很有用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号