当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Hierarchical Assembly of RNA Three-Dimensional Structures Based on Loop Templates.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-01-08 , DOI: 10.1021/acs.jpcb.7b10102 Xiaojun Xu 1, 2 , Shi-Jie Chen 2
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-01-08 , DOI: 10.1021/acs.jpcb.7b10102 Xiaojun Xu 1, 2 , Shi-Jie Chen 2
Affiliation
|
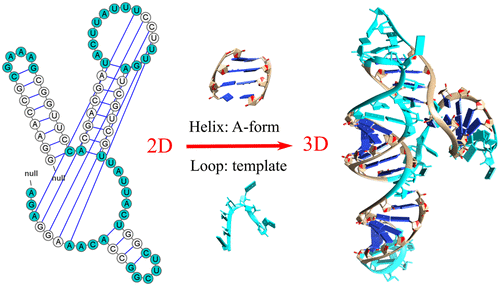
The current RNA structure prediction methods cannot keep up the pace of the rapidly increasing number of sequences and the emerging new functions of RNAs. Template-based RNA three-dimensional structure prediction methods are restricted by the limited number of known RNA structures, and traditional motif-based search for the templates does not always lead to successful results. Here we report a new template search and assembly algorithm, the hierarchical loop template-assembly method (VfoldLA). The method searches for templates for single strand loop/junctions instead of the whole motifs, which often renders no available templates, or short fragments (several nucleotides), which requires a long computational time to assemble and refine. The VfoldLA method has the advantage of accounting for local and nonlocal interloop interactions. Benchmark tests indicate that this new method can provide low-resolution predictions for RNA conformations at different levels of structural complexities. Furthermore, the VfoldLA-predicted conformations may also serve as reliable putative models for further structure prediction and refinements. VfoldLA is accessible at http://rna.physics.missouri.edu/vfoldLA .
中文翻译:

基于Loop模板的RNA三维结构的层次组装。
当前的RNA结构预测方法无法跟上快速增加的序列数量和新兴的RNA功能的步伐。基于模板的RNA三维结构预测方法受到有限数量的已知RNA结构的限制,并且传统的基于模板的模板搜索并不总是会导致成功的结果。在这里,我们报告了一种新的模板搜索和组装算法,即分层循环模板组装方法(VfoldLA)。该方法搜索单链环/接头而不是整个基序的模板,这通常无法提供可用的模板或较短的片段(几个核苷酸),这需要很长的计算时间才能组装和精炼。VfoldLA方法的优点是考虑了局部和非局部回路间的交互作用。基准测试表明,这种新方法可以为不同结构复杂性水平的RNA构象提供低分辨率的预测。此外,VfoldLA预测的构象也可用作进一步的结构预测和优化的可靠推定模型。可从http://rna.physics.missouri.edu/vfoldLA访问VfoldLA。
更新日期:2017-12-20
中文翻译:

基于Loop模板的RNA三维结构的层次组装。
当前的RNA结构预测方法无法跟上快速增加的序列数量和新兴的RNA功能的步伐。基于模板的RNA三维结构预测方法受到有限数量的已知RNA结构的限制,并且传统的基于模板的模板搜索并不总是会导致成功的结果。在这里,我们报告了一种新的模板搜索和组装算法,即分层循环模板组装方法(VfoldLA)。该方法搜索单链环/接头而不是整个基序的模板,这通常无法提供可用的模板或较短的片段(几个核苷酸),这需要很长的计算时间才能组装和精炼。VfoldLA方法的优点是考虑了局部和非局部回路间的交互作用。基准测试表明,这种新方法可以为不同结构复杂性水平的RNA构象提供低分辨率的预测。此外,VfoldLA预测的构象也可用作进一步的结构预测和优化的可靠推定模型。可从http://rna.physics.missouri.edu/vfoldLA访问VfoldLA。











































 京公网安备 11010802027423号
京公网安备 11010802027423号