当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
SmilesDrawer: Parsing and Drawing SMILES-Encoded Molecular Structures Using Client-Side JavaScript
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-11 00:00:00 , DOI: 10.1021/acs.jcim.7b00425 Daniel Probst 1 , Jean-Louis Reymond 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-01-11 00:00:00 , DOI: 10.1021/acs.jcim.7b00425 Daniel Probst 1 , Jean-Louis Reymond 1
Affiliation
|
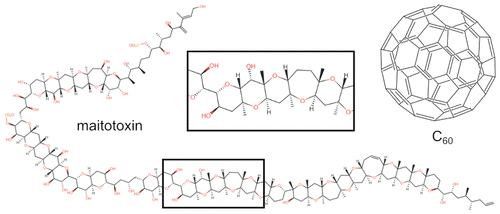
Here we present SmilesDrawer, a dependency-free JavaScript component capable of both parsing and drawing SMILES-encoded molecular structures client-side, developed to be easily integrated into web projects and to display organic molecules in large numbers and fast succession. SmilesDrawer can draw structurally and stereochemically complex structures such as maitotoxin and C60 without using templates, yet has an exceptionally small computational footprint and low memory usage without the requirement for loading images or any other form of client–server communication, making it easy to integrate even in secure (intranet, firewalled) or offline applications. These features allow the rendering of thousands of molecular structure drawings on a single web page within seconds on a wide range of hardware supporting modern browsers. The source code as well as the most recent build of SmilesDrawer is available on Github (http://doc.gdb.tools/smilesDrawer/). Both yarn and npm packages are also available.
中文翻译:

SmilesDrawer:使用客户端JavaScript解析和绘制SMILES编码的分子结构
在这里,我们介绍SmilesDrawer,这是一个无依赖的JavaScript组件,能够解析和绘制客户端SMILES编码的分子结构,其开发目的是易于集成到Web项目中,并能够快速连续显示大量有机分子。SmilesDrawer可以绘制结构和立体化学复杂的结构,例如麦托毒素和C 60不使用模板,却具有极小的计算空间和较低的内存使用量,而无需加载图像或任何其他形式的客户端-服务器通信,从而即使在安全(内部网,防火墙)或脱机应用程序中也可以轻松集成。这些功能允许在支持现代浏览器的各种硬件上在几秒钟内在单个网页上绘制数千个分子结构图。Github(http://doc.gdb.tools/smilesDrawer/)上提供了源代码以及最新版本的SmilesDrawer。也可以提供yarn和npm两种包装。
更新日期:2018-01-11
中文翻译:

SmilesDrawer:使用客户端JavaScript解析和绘制SMILES编码的分子结构
在这里,我们介绍SmilesDrawer,这是一个无依赖的JavaScript组件,能够解析和绘制客户端SMILES编码的分子结构,其开发目的是易于集成到Web项目中,并能够快速连续显示大量有机分子。SmilesDrawer可以绘制结构和立体化学复杂的结构,例如麦托毒素和C 60不使用模板,却具有极小的计算空间和较低的内存使用量,而无需加载图像或任何其他形式的客户端-服务器通信,从而即使在安全(内部网,防火墙)或脱机应用程序中也可以轻松集成。这些功能允许在支持现代浏览器的各种硬件上在几秒钟内在单个网页上绘制数千个分子结构图。Github(http://doc.gdb.tools/smilesDrawer/)上提供了源代码以及最新版本的SmilesDrawer。也可以提供yarn和npm两种包装。











































 京公网安备 11010802027423号
京公网安备 11010802027423号