当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
TARSyn: Tunable Antibiotic Resistance Devices Enabling Bacterial Synthetic Evolution and Protein Production
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-01-04 00:00:00 , DOI: 10.1021/acssynbio.7b00200 Maja Rennig 1 , Virginia Martinez 1 , Kiavash Mirzadeh 2, 3 , Finn Dunas 4 , Belinda Röjsäter 4 , Daniel O. Daley 2, 3 , Morten H. H. Nørholm 1, 3
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-01-04 00:00:00 , DOI: 10.1021/acssynbio.7b00200 Maja Rennig 1 , Virginia Martinez 1 , Kiavash Mirzadeh 2, 3 , Finn Dunas 4 , Belinda Röjsäter 4 , Daniel O. Daley 2, 3 , Morten H. H. Nørholm 1, 3
Affiliation
|
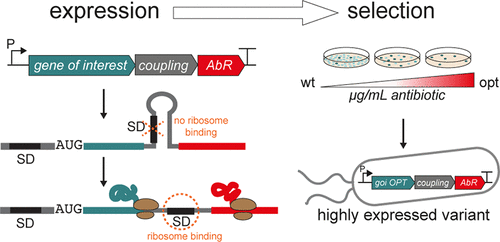
Evolution can be harnessed to optimize synthetic biology designs. A prominent example is recombinant protein production—a dominating theme in biotechnology for more than three decades. Typically, a protein coding sequence (cds) is recombined with genetic elements, such as promoters, ribosome binding sites and terminators, which control expression in a cell factory. A major bottleneck during production is translational initiation. Previously we identified more effective translation initiation regions (TIRs) by creating sequence libraries and then selecting for a TIR that drives high-level expression—an example of synthetic evolution. However, manual screening limits the ability to assay expression levels of all putative sequences in the libraries. Here we have solved this bottleneck by designing a collection of translational coupling devices based on a RNA secondary structure. Exchange of different sequence elements in this device allows for different coupling efficiencies, therefore giving the devices a tunable nature. Sandwiching these devices between the cds and an antibiotic selection marker that functions over a broad dynamic range of antibiotic concentrations adds to the tunability and allows expression levels in large clone libraries to be probed using a simple cell survival assay on the respective antibiotic. The power of the approach is demonstrated by substantially increasing production of two commercially interesting proteins, a Nanobody and an Affibody. The method is a simple and inexpensive alternative to advanced screening techniques that can be carried out in any laboratory.
中文翻译:

TARSyn:可调节的抗生素抗性设备,可实现细菌的合成进化和蛋白质生产
可以利用进化来优化合成生物学设计。一个突出的例子是重组蛋白生产,这是生物技术三十年来的主导主题。通常,蛋白质编码序列(cds)与遗传元件重组,例如控制细胞工厂中表达的启动子,核糖体结合位点和终止子。生产过程中的主要瓶颈是翻译启动。以前,我们通过创建序列库然后选择驱动高水平表达的TIR(合成进化的一个例子)来确定更有效的翻译起始区(TIR)。但是,手动筛选限制了分析文库中所有推定序列表达水平的能力。在这里,我们通过设计基于RNA二级结构的翻译偶联设备的集合解决了这一瓶颈。在该装置中交换不同的序列元件允许不同的偶联效率,因此赋予装置可调的性质。将这些设备夹在cds和在广泛的抗生素浓度动态范围内起作用的抗生素选择标记之间,增加了可调性,并允许使用简单的细胞存活测定法对相应的抗生素探测大型克隆文库中的表达水平。该方法的功能通过大幅增加两种商业上感兴趣的蛋白质(纳米抗体和亲和抗体)的生产来证明。该方法是可在任何实验室中进行的高级筛选技术的一种简单且廉价的替代方法。
更新日期:2018-01-04
中文翻译:

TARSyn:可调节的抗生素抗性设备,可实现细菌的合成进化和蛋白质生产
可以利用进化来优化合成生物学设计。一个突出的例子是重组蛋白生产,这是生物技术三十年来的主导主题。通常,蛋白质编码序列(cds)与遗传元件重组,例如控制细胞工厂中表达的启动子,核糖体结合位点和终止子。生产过程中的主要瓶颈是翻译启动。以前,我们通过创建序列库然后选择驱动高水平表达的TIR(合成进化的一个例子)来确定更有效的翻译起始区(TIR)。但是,手动筛选限制了分析文库中所有推定序列表达水平的能力。在这里,我们通过设计基于RNA二级结构的翻译偶联设备的集合解决了这一瓶颈。在该装置中交换不同的序列元件允许不同的偶联效率,因此赋予装置可调的性质。将这些设备夹在cds和在广泛的抗生素浓度动态范围内起作用的抗生素选择标记之间,增加了可调性,并允许使用简单的细胞存活测定法对相应的抗生素探测大型克隆文库中的表达水平。该方法的功能通过大幅增加两种商业上感兴趣的蛋白质(纳米抗体和亲和抗体)的生产来证明。该方法是可在任何实验室中进行的高级筛选技术的一种简单且廉价的替代方法。



























 京公网安备 11010802027423号
京公网安备 11010802027423号