Chemical Physics ( IF 2.0 ) Pub Date : 2017-12-19 , DOI: 10.1016/j.chemphys.2017.12.010 Gordon Hithell , Paul M. Donaldson , Gregory M. Greetham , Michael Towrie , Anthony W. Parker , Glenn A. Burley , Neil T. Hunt
|
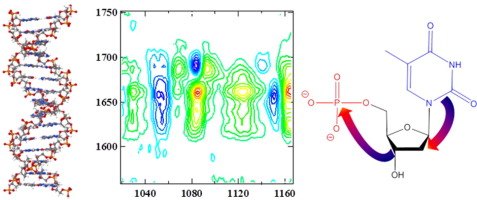
The effect of oligomer length on the vibrational mode coupling and energy relaxation mechanisms of AT-rich DNA oligomers in double- and single-stranded conformations has been investigated using two-dimensional infrared spectroscopy. Vibrational coupling of modes of the DNA bases to the symmetric stretching vibration of the backbone phosphate group was observed for oligomers long enough to form duplex-DNA structures. The coupling was lost upon melting of the duplex. No significant effect of oligomer length or DNA secondary structure was found on either the timescale for vibrational relaxation of the base modes or the mechanism, which was consistent with a cascade process from base modes to intermediate modes, some of which are located on the deoxyribose group, and subsequently to the phosphate backbone. The study shows that vibrational coupling between base and backbone requires formation of the double-helix structure while vibrational energy management is an inherent property of the nucleotide.
中文翻译:

低聚物长度对双链DNA振动耦合和能量弛豫的影响
使用二维红外光谱研究了低聚物长度对双链和单链构象中富含AT的DNA低聚物的振动模式耦合和能量弛豫机制的影响。对于足够长以形成双链DNA结构的寡聚物,观察到DNA碱基模式与骨架磷酸酯基团的对称拉伸振动的振动偶联。双链体融化后失去偶联。在基本模式振动弛豫的时间尺度上或机理上,均未发现寡聚物长度或DNA二级结构的显着影响,这与从基本模式到中间模式的级联过程一致,其中一些位于脱氧核糖基团上,然后是磷酸盐骨架。











































 京公网安备 11010802027423号
京公网安备 11010802027423号