当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
MinGenome: An In Silico Top-Down Approach for the Synthesis of Minimized Genomes
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-01-20 00:00:00 , DOI: 10.1021/acssynbio.7b00296 Lin Wang 1 , Costas D. Maranas 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-01-20 00:00:00 , DOI: 10.1021/acssynbio.7b00296 Lin Wang 1 , Costas D. Maranas 1
Affiliation
|
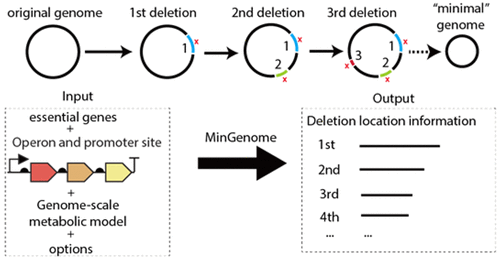
Genome minimized strains offer advantages as production chassis by reducing transcriptional cost, eliminating competing functions and limiting unwanted regulatory interactions. Existing approaches for identifying stretches of DNA to remove are largely ad hoc based on information on presumably dispensable regions through experimentally determined nonessential genes and comparative genomics. Here we introduce a versatile genome reduction algorithm MinGenome that implements a mixed-integer linear programming (MILP) algorithm to identify in size descending order all dispensable contiguous sequences without affecting the organism’s growth or other desirable traits. Known essential genes or genes that cause significant fitness or performance loss can be flagged and their deletion can be prohibited. MinGenome also preserves needed transcription factors and promoter regions ensuring that retained genes will be properly transcribed while also avoiding the simultaneous deletion of synthetic lethal pairs. The potential benefit of removing even larger contiguous stretches of DNA if only one or two essential genes (to be reinserted elsewhere) are within the deleted sequence is explored. We applied the algorithm to design a minimized E. coli strain and found that we were able to recapitulate the long deletions identified in previous experimental studies and discover alternative combinations of deletions that have not yet been explored in vivo.
中文翻译:

MinGenome:最小化基因组合成的计算机模拟自上而下方法
最小化基因组的菌株通过降低转录成本,消除竞争功能并限制有害的调控相互作用,提供了作为生产底盘的优势。识别要去除的DNA片段的现有方法在很大程度上是临时的基于通过实验确定的非必需基因和比较基因组学得出的大概可有可无区域的信息。在这里,我们介绍了一种通用的基因组缩减算法MinGenome,该算法实现了混合整数线性规划(MILP)算法,以按大小降序识别所有可分配的连续序列,而不会影响生物的生长或其他所需性状。可以标记已知的必需基因或会导致严重适应性或性能下降的基因,并禁止其删除。MinGenome还保留了所需的转录因子和启动子区域,以确保保留的基因将被正确转录,同时还避免了合成致死对的同时缺失。如果只有一个或两个必需基因(要在其他位置重新插入)位于删除的序列中,则探索去除更大的连续DNA片段的潜在益处。我们应用该算法设计了最小化大肠杆菌菌株,发现我们能够概括先前实验研究中鉴定的长缺失,并发现尚未在体内探索的缺失的替代组合。
更新日期:2018-01-20
中文翻译:

MinGenome:最小化基因组合成的计算机模拟自上而下方法
最小化基因组的菌株通过降低转录成本,消除竞争功能并限制有害的调控相互作用,提供了作为生产底盘的优势。识别要去除的DNA片段的现有方法在很大程度上是临时的基于通过实验确定的非必需基因和比较基因组学得出的大概可有可无区域的信息。在这里,我们介绍了一种通用的基因组缩减算法MinGenome,该算法实现了混合整数线性规划(MILP)算法,以按大小降序识别所有可分配的连续序列,而不会影响生物的生长或其他所需性状。可以标记已知的必需基因或会导致严重适应性或性能下降的基因,并禁止其删除。MinGenome还保留了所需的转录因子和启动子区域,以确保保留的基因将被正确转录,同时还避免了合成致死对的同时缺失。如果只有一个或两个必需基因(要在其他位置重新插入)位于删除的序列中,则探索去除更大的连续DNA片段的潜在益处。我们应用该算法设计了最小化大肠杆菌菌株,发现我们能够概括先前实验研究中鉴定的长缺失,并发现尚未在体内探索的缺失的替代组合。











































 京公网安备 11010802027423号
京公网安备 11010802027423号