当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Excitation energy transfer pathways in light-harvesting proteins: Modeling with PyFREC
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-12-15 , DOI: 10.1002/jcc.25134 Yana Kholod 1 , Michael DeFilippo 1 , Brittany Reed 1 , Danielle Valdez 1 , Grant Gillan 1 , Dmytro Kosenkov 1
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-12-15 , DOI: 10.1002/jcc.25134 Yana Kholod 1 , Michael DeFilippo 1 , Brittany Reed 1 , Danielle Valdez 1 , Grant Gillan 1 , Dmytro Kosenkov 1
Affiliation
|
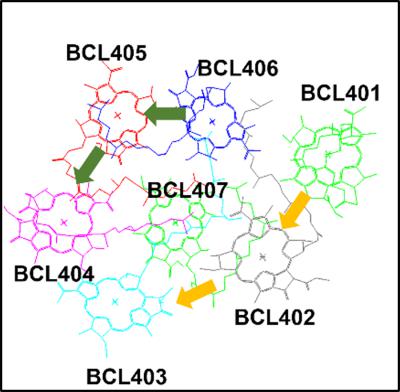
Excitation energy transfer (EET) determines the fate of sunlight energy absorbed by light‐harvesting proteins in natural photosynthetic systems and photovoltaic cells. As previously reported (D. Kosenkov, J. Comput. Chem. 2016, 37(19), 1847), PyFREC software enables computation of electronic couplings between organic molecules with a molecular fragmentation approach. The present work reports implementation of direct fragmentation‐based computation of the electronic couplings and EET rates in pigment–protein complexes within the Förster theory in PyFREC. The new feature enables assessment of EET pathways in a wide range of photosynthetic complexes, as well as artificial molecular architectures that include light‐harvesting proteins or tagged fluorescent biomolecules. The developed methodology has been tested analyzing EET in the Fenna–Matthews–Olson (FMO) pigment–protein complex. The pathways of excitation energy transfer in FMO have been identified based on the kinetics studies. © 2017 Wiley Periodicals, Inc.
中文翻译:

光捕获蛋白质中的激发能量转移途径:使用 PyFREC 建模
激发能量转移(EET)决定了自然光合作用系统和光伏电池中的光捕获蛋白吸收的阳光能量的命运。正如之前报道的那样(D. Kosenkov, J. Comput. Chem. 2016, 37(19), 1847),PyFREC 软件能够通过分子碎片化方法计算有机分子之间的电子耦合。目前的工作报告了 PyFREC 中 Förster 理论中色素-蛋白质复合物中电子耦合和 EET 速率的直接碎片化计算的实现。新功能可以评估各种光合复合物中的 EET 途径,以及包括光捕获蛋白或标记荧光生物分子的人工分子结构。所开发的方法已经在 Fenna-Matthews-Olson (FMO) 色素-蛋白质复合物中分析 EET 进行了测试。基于动力学研究已经确定了 FMO 中激发能量转移的途径。© 2017 威利期刊公司。
更新日期:2017-12-15
中文翻译:

光捕获蛋白质中的激发能量转移途径:使用 PyFREC 建模
激发能量转移(EET)决定了自然光合作用系统和光伏电池中的光捕获蛋白吸收的阳光能量的命运。正如之前报道的那样(D. Kosenkov, J. Comput. Chem. 2016, 37(19), 1847),PyFREC 软件能够通过分子碎片化方法计算有机分子之间的电子耦合。目前的工作报告了 PyFREC 中 Förster 理论中色素-蛋白质复合物中电子耦合和 EET 速率的直接碎片化计算的实现。新功能可以评估各种光合复合物中的 EET 途径,以及包括光捕获蛋白或标记荧光生物分子的人工分子结构。所开发的方法已经在 Fenna-Matthews-Olson (FMO) 色素-蛋白质复合物中分析 EET 进行了测试。基于动力学研究已经确定了 FMO 中激发能量转移的途径。© 2017 威利期刊公司。











































 京公网安备 11010802027423号
京公网安备 11010802027423号