当前位置:
X-MOL 学术
›
ACS Chem. Neurosci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The Associative Memory, Water Mediated, Structure and Energy Model (AWSEM)-Amylometer: Predicting Amyloid Propensity and Fibril Topology Using an Optimized Folding Landscape Model
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2017-12-14 00:00:00 , DOI: 10.1021/acschemneuro.7b00436 Mingchen Chen , Nicholas P. Schafer , Weihua Zheng , Peter G. Wolynes
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2017-12-14 00:00:00 , DOI: 10.1021/acschemneuro.7b00436 Mingchen Chen , Nicholas P. Schafer , Weihua Zheng , Peter G. Wolynes
|
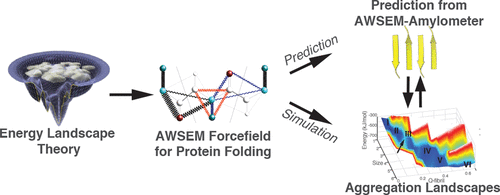
Amyloids are fibrillar protein aggregates with simple repeated structural motifs in their cores, usually β-strands but sometimes α-helices. Identifying the amyloid-prone regions within protein sequences is important both for understanding the mechanisms of amyloid-associated diseases and for understanding functional amyloids. Based on the crystal structures of seven cross-β amyloidogenic peptides with different topologies and one recently solved cross-α fiber structure, we have developed a computational approach for identifying amyloidogenic segments in protein sequences using the Associative memory, Water mediated, Structure and Energy Model (AWSEM). The AWSEM-Amylometer performs favorably in comparison with other predictors in predicting aggregation-prone sequences in multiple data sets. The method also predicts well the specific topologies (the relative arrangement of β-strands in the core) of the amyloid fibrils. An important advantage of the AWSEM-Amylometer over other existing methods is its direct connection with an efficient, optimized protein folding simulation model, AWSEM. This connection allows one to combine efficient and accurate search of protein sequences for amyloidogenic segments with the detailed study of the thermodynamic and kinetic roles that these segments play in folding and aggregation in the context of the entire protein sequence. We present new simulation results that highlight the free energy landscapes of peptides that can take on multiple fibril topologies. We also demonstrate how the Amylometer methodology can be straightforwardly extended to the study of functional amyloids that have the recently discovered cross-α fibril architecture.
中文翻译:

联想记忆,水介导,结构和能量模型(AWSEM)-测速计:使用优化的折叠景观模型预测淀粉样蛋白的倾向和原纤维拓扑
淀粉样蛋白是纤维状蛋白聚集体,其核心中具有简单的重复结构基序,通常为β链,但有时为α螺旋。鉴定蛋白质序列中易发生淀粉样蛋白的区域对于理解淀粉样蛋白相关疾病的机制和理解功能性淀粉样蛋白都是重要的。基于七个具有不同拓扑结构的交叉β淀粉样蛋白生成肽的晶体结构和一个最近解决的交叉α纤维结构,我们开发了一种使用关联记忆,水介导,结构和能量模型鉴定蛋白质序列中淀粉样蛋白生成片段的计算方法。 (AWSEM)。与其他预测变量相比,AWSEM测速仪在预测多个数据集中易于聚集的序列时表现出色。该方法还可以很好地预测淀粉样蛋白原纤维的特定拓扑结构(核心中β链的相对排列)。与其他现有方法相比,AWSEM测速仪的重要优势在于它与高效,优化的蛋白质折叠模拟模型AWSEM的直接连接。这种联系使人们可以结合有效,准确地搜索淀粉样蛋白生成片段的蛋白质序列,以及对这些片段在整个蛋白质序列中在折叠和聚集中发挥的热力学和动力学作用的详细研究。我们提出了新的模拟结果,突出了可以具有多种原纤维拓扑结构的肽的自由能态。
更新日期:2017-12-14
中文翻译:

联想记忆,水介导,结构和能量模型(AWSEM)-测速计:使用优化的折叠景观模型预测淀粉样蛋白的倾向和原纤维拓扑
淀粉样蛋白是纤维状蛋白聚集体,其核心中具有简单的重复结构基序,通常为β链,但有时为α螺旋。鉴定蛋白质序列中易发生淀粉样蛋白的区域对于理解淀粉样蛋白相关疾病的机制和理解功能性淀粉样蛋白都是重要的。基于七个具有不同拓扑结构的交叉β淀粉样蛋白生成肽的晶体结构和一个最近解决的交叉α纤维结构,我们开发了一种使用关联记忆,水介导,结构和能量模型鉴定蛋白质序列中淀粉样蛋白生成片段的计算方法。 (AWSEM)。与其他预测变量相比,AWSEM测速仪在预测多个数据集中易于聚集的序列时表现出色。该方法还可以很好地预测淀粉样蛋白原纤维的特定拓扑结构(核心中β链的相对排列)。与其他现有方法相比,AWSEM测速仪的重要优势在于它与高效,优化的蛋白质折叠模拟模型AWSEM的直接连接。这种联系使人们可以结合有效,准确地搜索淀粉样蛋白生成片段的蛋白质序列,以及对这些片段在整个蛋白质序列中在折叠和聚集中发挥的热力学和动力学作用的详细研究。我们提出了新的模拟结果,突出了可以具有多种原纤维拓扑结构的肽的自由能态。











































 京公网安备 11010802027423号
京公网安备 11010802027423号