Journal of Inorganic Biochemistry ( IF 3.8 ) Pub Date : 2017-12-13 , DOI: 10.1016/j.jinorgbio.2017.12.008 Stephen M. Keable , Jacopo Vertemara , Oleg A. Zadvornyy , Brian J. Eilers , Karamatullah Danyal , Andrew J. Rasmussen , Luca De Gioia , Giuseppe Zampella , Lance C. Seefeldt , John W. Peters
|
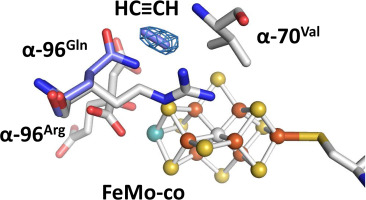
The biological reduction of dinitrogen (N2) to ammonia is catalyzed by the complex metalloenzyme nitrogenase. Structures of the nitrogenase component proteins, Iron (Fe) protein and Molybdenum‑iron (MoFe) protein, and the stabilized complexes these component proteins, have been determined, providing a foundation for a number of fundamental aspects of the complicated catalytic mechanism. The reduction of dinitrogen to ammonia is a complex process that involves the binding of N2 followed by reduction with multiple electrons and protons. Electron transfer into nitrogenase is typically constrained to the unique electron donor, the Fe protein. These constraints have prevented structural characterization of the active site with bound substrate. Recently it has been realized that selected amino acid substitutions in the environment of the active site metal cluster (Iron‑molybdenum cofactor, FeMo-co) allow substrates to persist even in the resting state. Reported here is a 1.70 Å crystal structure of a nitrogenase MoFe protein α-96Arg ➔ Gln variant with the alternative substrate acetylene trapped in a channel in close proximity to FeMo-co. Complementary theoretical calculations support the validity of the acetylene interaction at this site and is also consistent with more favorable interactions in the variant MoFe protein compared to the native MoFe protein. This work represents the first structural evidence of a substrate trapped in the nitrogenase MoFe protein and is consistent with earlier assignments of proposed substrate pathways and substrate binding sites deduced from biochemical, spectroscopic, and theoretical studies.
中文翻译:

底物乙炔被困在活性位点附近的固氮酶钼铁蛋白的结构表征
复杂的金属酶固氮酶催化将二氮(N 2)生物还原为氨。已确定了固氮酶成分蛋白,铁(Fe)蛋白和钼铁(MoFe)蛋白的结构,以及这些成分蛋白的稳定复合物,为复杂的催化机理的许多基本方面提供了基础。将二氮还原为氨是一个复杂的过程,涉及N 2的结合然后用多个电子和质子还原。电子转移到固氮酶中通常受限于唯一的电子供体Fe蛋白。这些限制阻止了具有结合的底物的活性位点的结构表征。最近,人们已经认识到,在活性位点金属簇(铁-钼辅因子,FeMo-co)的环境中选择的氨基酸取代可使底物即使在静止状态下也能持久存在。这里报告是固氮钼铁蛋白的1.70的晶体结构α-96精氨酸➔谷氨酰胺变体,乙炔的另一种底物被困在紧邻FeMo-co的通道中。互补的理论计算支持该部位乙炔相互作用的有效性,并且与天然MoFe蛋白相比,也与变异的MoFe蛋白中更有利的相互作用相一致。这项工作代表了被困在固氮酶MoFe蛋白中的底物的第一个结构证据,并且与从生物化学,光谱学和理论研究推论得出的底物途径和底物结合位点的早期分配相一致。











































 京公网安备 11010802027423号
京公网安备 11010802027423号