当前位置:
X-MOL 学术
›
Biomater. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Shape-specific nanostructured protein mimics from de novo designed chimeric peptides†
Biomaterials Science ( IF 5.8 ) Pub Date : 2017-12-11 00:00:00 , DOI: 10.1039/c7bm00906b Linhai Jiang 1, 2, 3, 4 , Su Yang 1, 2, 3, 4 , Reidar Lund 5, 6, 7, 8 , He Dong 1, 2, 3, 4
Biomaterials Science ( IF 5.8 ) Pub Date : 2017-12-11 00:00:00 , DOI: 10.1039/c7bm00906b Linhai Jiang 1, 2, 3, 4 , Su Yang 1, 2, 3, 4 , Reidar Lund 5, 6, 7, 8 , He Dong 1, 2, 3, 4
Affiliation
|
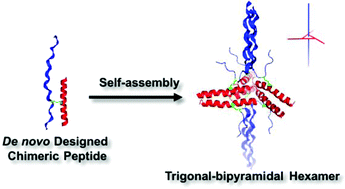
Natural proteins self-assemble into highly-ordered nanoscaled architectures to perform specific functions. The intricate functions of proteins have provided great impetus for researchers to develop strategies for designing and engineering synthetic nanostructures as protein mimics. Compared to the success in engineering fibrous protein mimetics, the design of discrete globular protein-like nanostructures has been challenging mainly due to the lack of precise control over geometric packing and intermolecular interactions among synthetic building blocks. In this contribution, we report an effective strategy to construct shape-specific nanostructures based on the self-assembly of chimeric peptides consisting of a coiled coil dimer and a collagen triple helix folding motif. Under salt-free conditions, we showed spontaneous self-assembly of the chimeric peptides into monodisperse, trigonal bipyramidal-like nanoparticles with precise control over the stoichiometry of two folding motifs and the geometrical arrangements relative to one another. Three coiled coil dimers are interdigitated on the equatorial plane while the two collagen triple helices are located in the axial position, perpendicular to the coiled coil plane. A detailed molecular model was proposed and further validated by small angle X-ray scattering experiments and molecular dynamics (MD) simulation. The results from this study indicated that the molecular folding of each motif within the chimeric peptides and their geometric packing played important roles in the formation of discrete protein-like nanoparticles. The peptide design and self-assembly mechanism may open up new routes for the construction of highly organized, discrete self-assembling protein-like nanostructures with greater levels of control over assembly accuracy.
中文翻译:

从头设计的嵌合肽 模仿形状特定的纳米结构蛋白质†
天然蛋白质自组装成高度有序的纳米级结构,以执行特定功能。蛋白质的复杂功能为研究人员开发作为蛋白质模拟物的设计和工程化合成纳米结构的策略提供了强大的动力。与工程化纤维蛋白模拟物的成功相比,离散球状蛋白状纳米结构的设计具有挑战性,这主要是由于缺乏对几何堆积和合成构件之间的分子间相互作用的精确控制所致。在这项贡献中,我们报告了一种有效的策略,可基于嵌合肽的自组装构建形状特定的纳米结构,所述嵌合肽由卷曲的螺旋二聚体和胶原三螺旋折叠基序组成。在无盐条件下,我们展示了嵌合肽的自发自组装成单分散的,三角形的双锥体状纳米颗粒,并精确控制了两个折叠基序的化学计量以及相对于彼此的几何排列。三个盘绕的二聚体在赤道平面上相互交叉,而两个胶原三螺旋位于轴向位置,垂直于盘绕的螺旋平面。提出了详细的分子模型,并通过小角度X射线散射实验和分子动力学(MD)模拟进一步验证了该模型。这项研究的结果表明,嵌合肽中每个基序的分子折叠及其几何堆积在离散蛋白样纳米颗粒的形成中起着重要作用。
更新日期:2017-12-11
中文翻译:

从头设计的嵌合肽 模仿形状特定的纳米结构蛋白质†
天然蛋白质自组装成高度有序的纳米级结构,以执行特定功能。蛋白质的复杂功能为研究人员开发作为蛋白质模拟物的设计和工程化合成纳米结构的策略提供了强大的动力。与工程化纤维蛋白模拟物的成功相比,离散球状蛋白状纳米结构的设计具有挑战性,这主要是由于缺乏对几何堆积和合成构件之间的分子间相互作用的精确控制所致。在这项贡献中,我们报告了一种有效的策略,可基于嵌合肽的自组装构建形状特定的纳米结构,所述嵌合肽由卷曲的螺旋二聚体和胶原三螺旋折叠基序组成。在无盐条件下,我们展示了嵌合肽的自发自组装成单分散的,三角形的双锥体状纳米颗粒,并精确控制了两个折叠基序的化学计量以及相对于彼此的几何排列。三个盘绕的二聚体在赤道平面上相互交叉,而两个胶原三螺旋位于轴向位置,垂直于盘绕的螺旋平面。提出了详细的分子模型,并通过小角度X射线散射实验和分子动力学(MD)模拟进一步验证了该模型。这项研究的结果表明,嵌合肽中每个基序的分子折叠及其几何堆积在离散蛋白样纳米颗粒的形成中起着重要作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号