当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Robust Molecular Network Motif for Period-Doubling Devices
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2017-12-11 00:00:00 , DOI: 10.1021/acssynbio.7b00222 Christian Cuba Samaniego 1 , Elisa Franco 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2017-12-11 00:00:00 , DOI: 10.1021/acssynbio.7b00222 Christian Cuba Samaniego 1 , Elisa Franco 1
Affiliation
|
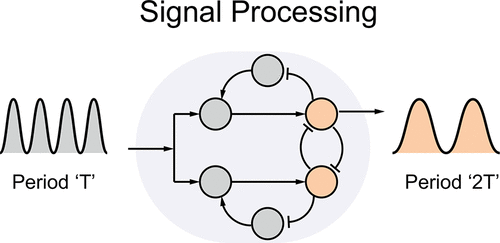
Life is sustained by a variety of cyclic processes such as cell division, muscle contraction, and neuron firing. The periodic signals powering these processes often direct a variety of other downstream systems, which operate at different time scales and must have the capacity to divide or multiply the period of the master clock. Period modulation is also an important challenge in synthetic molecular systems, where slow and fast components may have to be coordinated simultaneously by a single oscillator whose frequency is often difficult to tune. Circuits that can multiply the period of a clock signal (frequency dividers), such as binary counters and flip-flops, are commonly encountered in electronic systems, but design principles to obtain similar devices in biological systems are still unclear. We take inspiration from the architecture of electronic flip-flops, and we propose to build biomolecular period-doubling networks by combining a bistable switch with negative feedback modules that preprocess the circuit inputs. We identify a network motif and we show it can be “realized” using different biomolecular components; two of the realizations we propose rely on transcriptional gene networks and one on nucleic acid strand displacement systems. We examine the capacity of each realization to perform period-doubling by studying how bistability of the motif is affected by the presence of the input; for this purpose, we employ mathematical tools from algebraic geometry that provide us with valuable insights on the input/output behavior as a function of the realization parameters. We show that transcriptional network realizations operate correctly also in a stochastic regime when processing oscillations from the repressilator, a canonical synthetic in vivo oscillator. Finally, we compare the performance of different realizations in a range of realistic parameters via numerical sensitivity analysis of the period-doubling region, computed with respect to the input period and amplitude. Our mathematical and computational analysis suggests that the motif we propose is generally robust with respect to specific implementation details: functionally equivalent circuits can be built as long as the species-interaction topology is respected. This indicates that experimental construction of the circuit is possible with a variety of components within the rapidly expanding libraries available in synthetic biology.
中文翻译:

倍频设备的鲁棒分子网络图形
生命可以通过多种循环过程维持,例如细胞分裂,肌肉收缩和神经元放电。为这些过程提供动力的周期性信号通常会引导其他各种下游系统,这些系统在不同的时标下运行,并且必须具有划分或倍增主时钟周期的能力。在合成分子系统中,周期调制也是一个重要的挑战,在该合成分子系统中,慢速和快速分量可能必须由一个频率通常难以调节的单个振荡器同时进行协调。在电子系统中通常会遇到可倍增时钟信号周期(分频器)的电路,例如二进制计数器和触发器,但是在生物系统中获得类似器件的设计原理仍不清楚。我们从电子触发器的体系结构中汲取灵感,并建议通过将双稳态开关与负反馈模块结合使用来构建生物分子周期倍增网络,该模块对电路输入进行预处理。我们确定了一个网络主题,并表明可以使用不同的生物分子成分来“实现”它。我们提出的两种实现依赖于转录基因网络,一种依赖于核酸链置换系统。通过研究图案的双稳态如何受输入的存在影响,我们研究了每种实现方式执行周期倍增的能力。为此,我们采用了代数几何的数学工具,这些工具为我们提供了有关实现参数的输入/输出行为的宝贵见解。体内振荡器。最后,我们通过对周期倍增区域的数值敏感性分析(相对于输入周期和幅度进行计算),在一系列实际参数中比较了不同实现的性能。我们的数学和计算分析表明,相对于特定的实现细节,我们提出的主题通常是可靠的:只要遵守物种交互拓扑,就可以构建功能等效的电路。这表明,在合成生物学中迅速扩展的库中,利用各种组件进行电路的实验性构建是可能的。
更新日期:2017-12-11
中文翻译:

倍频设备的鲁棒分子网络图形
生命可以通过多种循环过程维持,例如细胞分裂,肌肉收缩和神经元放电。为这些过程提供动力的周期性信号通常会引导其他各种下游系统,这些系统在不同的时标下运行,并且必须具有划分或倍增主时钟周期的能力。在合成分子系统中,周期调制也是一个重要的挑战,在该合成分子系统中,慢速和快速分量可能必须由一个频率通常难以调节的单个振荡器同时进行协调。在电子系统中通常会遇到可倍增时钟信号周期(分频器)的电路,例如二进制计数器和触发器,但是在生物系统中获得类似器件的设计原理仍不清楚。我们从电子触发器的体系结构中汲取灵感,并建议通过将双稳态开关与负反馈模块结合使用来构建生物分子周期倍增网络,该模块对电路输入进行预处理。我们确定了一个网络主题,并表明可以使用不同的生物分子成分来“实现”它。我们提出的两种实现依赖于转录基因网络,一种依赖于核酸链置换系统。通过研究图案的双稳态如何受输入的存在影响,我们研究了每种实现方式执行周期倍增的能力。为此,我们采用了代数几何的数学工具,这些工具为我们提供了有关实现参数的输入/输出行为的宝贵见解。体内振荡器。最后,我们通过对周期倍增区域的数值敏感性分析(相对于输入周期和幅度进行计算),在一系列实际参数中比较了不同实现的性能。我们的数学和计算分析表明,相对于特定的实现细节,我们提出的主题通常是可靠的:只要遵守物种交互拓扑,就可以构建功能等效的电路。这表明,在合成生物学中迅速扩展的库中,利用各种组件进行电路的实验性构建是可能的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号