当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Statistical Coupling Analysis-Guided Library Design for the Discovery of Mutant Luciferases
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-28 00:00:00 , DOI: 10.1021/acs.biochem.7b01014 Mira D Liu 1 , Elliot A Warner 1 , Charlotte E Morrissey 1 , Caitlyn W Fick 1 , Taia S Wu 1 , Marya Y Ornelas 1 , Gabriela V Ochoa 1 , Brendan S Zhang , Colin M Rathbun , William B Porterfield , Jennifer A Prescher , Aaron M Leconte 1
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-28 00:00:00 , DOI: 10.1021/acs.biochem.7b01014 Mira D Liu 1 , Elliot A Warner 1 , Charlotte E Morrissey 1 , Caitlyn W Fick 1 , Taia S Wu 1 , Marya Y Ornelas 1 , Gabriela V Ochoa 1 , Brendan S Zhang , Colin M Rathbun , William B Porterfield , Jennifer A Prescher , Aaron M Leconte 1
Affiliation
|
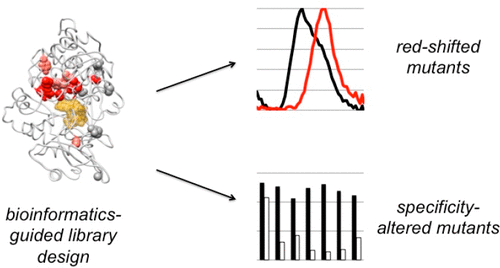
Directed evolution has proven to be an invaluable tool for protein engineering; however, there is still a need for developing new approaches to continue to improve the efficiency and efficacy of these methods. Here, we demonstrate a new method for library design that applies a previously developed bioinformatic method, Statistical Coupling Analysis (SCA). SCA uses homologous enzymes to identify amino acid positions that are mutable and functionally important and engage in synergistic interactions between amino acids. We use SCA to guide a library of the protein luciferase and demonstrate that, in a single round of selection, we can identify luciferase mutants with several valuable properties. Specifically, we identify luciferase mutants that possess both red-shifted emission spectra and improved stability relative to those of the wild-type enzyme. We also identify luciferase mutants that possess a >50-fold change in specificity for modified luciferins. To understand the mutational origin of these improved mutants, we demonstrate the role of mutations at N229, S239, and G246 in altered function. These studies show that SCA can be used to guide library design and rapidly identify synergistic amino acid mutations from a small library.
中文翻译:

用于发现突变荧光素酶的统计耦合分析引导文库设计
定向进化已被证明是蛋白质工程的宝贵工具。然而,仍然需要开发新的方法来继续提高这些方法的效率和功效。在这里,我们展示了一种新的文库设计方法,该方法应用了先前开发的生物信息学方法,即统计耦合分析(SCA)。 SCA 使用同源酶来识别可变且功能重要的氨基酸位置,并参与氨基酸之间的协同相互作用。我们使用 SCA 来指导蛋白质荧光素酶文库,并证明,在一轮选择中,我们可以识别具有多种有价值特性的荧光素酶突变体。具体来说,我们鉴定了相对于野生型酶具有红移发射光谱和改进的稳定性的荧光素酶突变体。我们还鉴定了对修饰荧光素的特异性具有 >50 倍变化的荧光素酶突变体。为了了解这些改良突变体的突变起源,我们证明了 N229、S239 和 G246 突变在功能改变中的作用。这些研究表明,SCA 可用于指导文库设计并快速识别小型文库中的协同氨基酸突变。
更新日期:2017-12-28
中文翻译:

用于发现突变荧光素酶的统计耦合分析引导文库设计
定向进化已被证明是蛋白质工程的宝贵工具。然而,仍然需要开发新的方法来继续提高这些方法的效率和功效。在这里,我们展示了一种新的文库设计方法,该方法应用了先前开发的生物信息学方法,即统计耦合分析(SCA)。 SCA 使用同源酶来识别可变且功能重要的氨基酸位置,并参与氨基酸之间的协同相互作用。我们使用 SCA 来指导蛋白质荧光素酶文库,并证明,在一轮选择中,我们可以识别具有多种有价值特性的荧光素酶突变体。具体来说,我们鉴定了相对于野生型酶具有红移发射光谱和改进的稳定性的荧光素酶突变体。我们还鉴定了对修饰荧光素的特异性具有 >50 倍变化的荧光素酶突变体。为了了解这些改良突变体的突变起源,我们证明了 N229、S239 和 G246 突变在功能改变中的作用。这些研究表明,SCA 可用于指导文库设计并快速识别小型文库中的协同氨基酸突变。











































 京公网安备 11010802027423号
京公网安备 11010802027423号