当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-19 00:00:00 , DOI: 10.1021/acs.biochem.7b01152 Sarah E. Jones 1 , Lars Olsen 1 , Michael Gajhede 1
Biochemistry ( IF 2.9 ) Pub Date : 2017-12-19 00:00:00 , DOI: 10.1021/acs.biochem.7b01152 Sarah E. Jones 1 , Lars Olsen 1 , Michael Gajhede 1
Affiliation
|
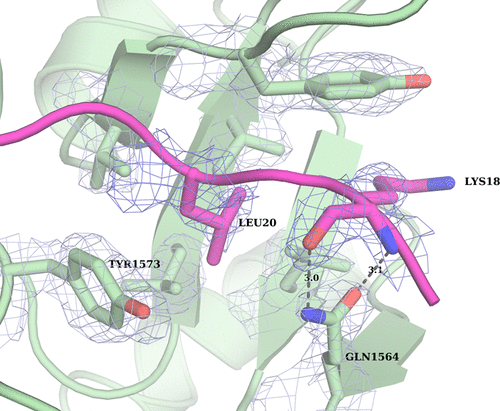
KDM subfamily 6 enzymes KDM6A and KDM6B specifically catalyze demethylation of di- and trimethylated lysine on histone 3 lysine 27 (H3K27me3/2) and play an important role in repression of developmental genes. Despite identical amino acid sequence in the immediate surroundings of H3K9me3/2 (ARKS), the enzymes do not catalyze demethylation of this general marker of repression. To address this question for KDM6B, we used computational methods to identify H3(17–33)-derived peptides with improved binding affinity that would allow co-crystallization with the catalytic core of human KDM6B (ccKDM6B). A total of five peptides were identified, and their IC50 values were determined in a matrix-assisted laser desorption ionization time-of-flight-based assay. Despite none of the peptides showing affinity significantly higher than that of the H3(17–33) peptide, it was possible to co-crystallize ccKDM6B with a H3(17–33)A21M peptide. This structure reveals the interactions between the KDM6B zinc binding domain and the H3(17–23) region. Although KDM6A and KDM6B differ in primary sequence, particularly in the H3L20 binding pocket of the zinc binding domains, where two histidines in KDM6A have been replaced by a glutamate and a tyrosine, they bind H3(17–23) in a very similar fashion. This structure shows that KDM6B, in analogy with KDM6A, also uses the zinc binding domain to achieve H3K27me3/me2 specificity. The histidine to glutamine substitution at amino acid position 1564 in the KDM6B zinc binding domain can further explain why KDM6B binds substrates with an affinity higher than that of KDM6A.
中文翻译:

组蛋白去甲基化酶KDM6B组蛋白3赖氨酸27特异性的结构基础
KDM亚家族6酶KDM6A和KDM6B特异性催化组蛋白3赖氨酸27(H3K27me3 / 2)上的二甲基和三甲基赖氨酸的去甲基化,并且在抑制发育基因中起重要作用。尽管在H3K9me3 / 2(AR K S)的周围环境中有相同的氨基酸序列,但这些酶并未催化这种一般性抑制标记的去甲基化。为了解决有关KDM6B的问题,我们使用了计算方法来鉴定具有改进的结合亲和力的H3(17-33)衍生肽,从而可以与人KDM6B(ccKDM6B)的催化核心共结晶。总共鉴定出五种肽,它们的IC 50在基于基质的激光解吸电离飞行时间测定中确定该值。尽管没有一种肽的亲和力明显高于H3(17-33)肽,但仍可以将ccKDM6B与H3(17-33)A21M肽共结晶。这种结构揭示了KDM6B锌结合域与H3(17-23)区之间的相互作用。尽管KDM6A和KDM6B的一级序列不同,尤其是锌结合域的H3L20结合口袋(其中KDM6A中的两个组氨酸已被谷氨酸和酪氨酸取代),但它们以非常相似的方式结合H3(17-23)。该结构表明,KDM6B与KDM6A类似,也使用锌结合域来实现H3K27me3 / me2特异性。
更新日期:2017-12-19
中文翻译:

组蛋白去甲基化酶KDM6B组蛋白3赖氨酸27特异性的结构基础
KDM亚家族6酶KDM6A和KDM6B特异性催化组蛋白3赖氨酸27(H3K27me3 / 2)上的二甲基和三甲基赖氨酸的去甲基化,并且在抑制发育基因中起重要作用。尽管在H3K9me3 / 2(AR K S)的周围环境中有相同的氨基酸序列,但这些酶并未催化这种一般性抑制标记的去甲基化。为了解决有关KDM6B的问题,我们使用了计算方法来鉴定具有改进的结合亲和力的H3(17-33)衍生肽,从而可以与人KDM6B(ccKDM6B)的催化核心共结晶。总共鉴定出五种肽,它们的IC 50在基于基质的激光解吸电离飞行时间测定中确定该值。尽管没有一种肽的亲和力明显高于H3(17-33)肽,但仍可以将ccKDM6B与H3(17-33)A21M肽共结晶。这种结构揭示了KDM6B锌结合域与H3(17-23)区之间的相互作用。尽管KDM6A和KDM6B的一级序列不同,尤其是锌结合域的H3L20结合口袋(其中KDM6A中的两个组氨酸已被谷氨酸和酪氨酸取代),但它们以非常相似的方式结合H3(17-23)。该结构表明,KDM6B与KDM6A类似,也使用锌结合域来实现H3K27me3 / me2特异性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号