Molecular Cell ( IF 14.5 ) Pub Date : 2017-12-07 , DOI: 10.1016/j.molcel.2017.11.014 Michal Rabani , Lindsey Pieper , Guo-Liang Chew , Alexander F. Schier
|
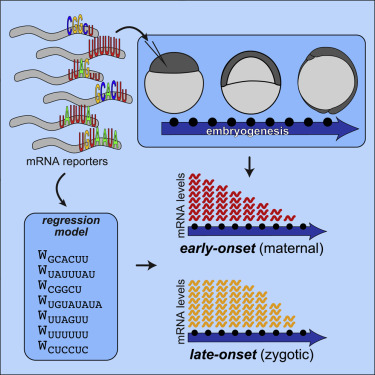
The stability of mRNAs is regulated by signals within their sequences, but a systematic and predictive understanding of the underlying sequence rules remains elusive. Here we introduce UTR-seq, a combination of massively parallel reporter assays and regression models, to survey the dynamics of tens of thousands of 3′ UTR sequences during early zebrafish embryogenesis. UTR-seq revealed two temporal degradation programs: a maternally encoded early-onset program and a late-onset program that accelerated degradation after zygotic genome activation. Three signals regulated early-onset rates: stabilizing poly-U and UUAG sequences and destabilizing GC-rich signals. Three signals explained late-onset degradation: miR-430 seeds, AU-rich sequences, and Pumilio recognition sites. Sequence-based regression models translated 3′ UTRs into their unique decay patterns and predicted the in vivo effect of sequence signals on mRNA stability. Their application led to the successful design of artificial 3′ UTRs that conferred specific mRNA dynamics. UTR-seq provides a general strategy to uncover the rules of RNA cis regulation.
中文翻译:

3'UTR序列的大规模平行记者分析确定了mRNA降解的体内规则。
mRNA的稳定性受其序列内信号的调节,但是对潜在序列规则的系统性和预测性了解仍然难以捉摸。在这里,我们介绍了UTR-seq,它是大规模并行报道基因分析和回归模型的组合,用于调查斑马鱼早期胚胎发生过程中数以万计的3'UTR序列的动力学。UTR-seq揭示了两个时间降解程序:一个母本编码的早期发作程序和一个晚期突变程序,它们在合子基因组激活后加速了降解。三种信号调节了早期发病率:稳定多聚U和UUAG序列和稳定富含GC的信号。三个信号解释了迟发性降解:miR-430种子,富含AU的序列和Pumilio识别位点。序列信号在体内对mRNA稳定性的影响。他们的应用导致成功设计了赋予特定mRNA动态的人工3'UTR。UTR-seq提供了揭示RNA顺式调控规则的一般策略。











































 京公网安备 11010802027423号
京公网安备 11010802027423号