Molecular Cell ( IF 14.5 ) Pub Date : 2017-12-07 , DOI: 10.1016/j.molcel.2017.11.015 Srinivas Ramachandran 1 , Kami Ahmad 2 , Steven Henikoff 1
|
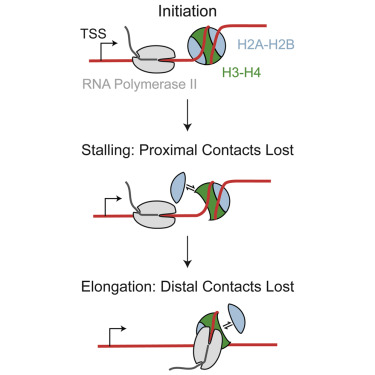
Nucleosomes are disrupted during transcription and other active processes, but the structural intermediates during nucleosome disruption in vivo are unknown. To identify intermediates, we mapped subnucleosomal protections in Drosophila cells using Micrococcal Nuclease followed by sequencing. At the first nucleosome position downstream of the transcription start site, we identified unwrapped intermediates, including hexasomes that lack either proximal or distal contacts. Inhibiting topoisomerases or depleting histone chaperones increased unwrapping, whereas inhibiting release of paused RNAPII or reducing RNAPII elongation decreased unwrapping. Our results indicate that positive torsion generated by elongating RNAPII causes transient loss of histone-DNA contacts. Using this mapping approach, we found that nucleosomes flanking human CTCF insulation sites are similarly disrupted. We also identified diagnostic subnucleosomal particle remnants in cell-free human DNA data as a relic of transcribed genes from apoptosing cells. Thus identification of subnucleosomal fragments from nuclease protection data represents a general strategy for structural epigenomics.
中文翻译:

转录和重塑产生不对称解包的核小体中间体
核小体在转录和其他活性过程中被破坏,但体内核小体破坏过程中的结构中间体尚不清楚。为了鉴定中间体,我们使用微球菌核酸酶绘制了果蝇细胞中的亚核小体保护图,然后进行测序。在转录起始位点下游的第一个核小体位置,我们鉴定了未包裹的中间体,包括缺乏近端或远端接触的六体。抑制拓扑异构酶或消耗组蛋白伴侣会增加解包,而抑制暂停的 RNAPII 的释放或减少 RNAPII 延伸会减少解包。我们的结果表明,延长 RNAPII 产生的正扭转会导致组蛋白-DNA 接触的短暂损失。使用这种作图方法,我们发现人类 CTCF 绝缘位点侧翼的核小体也受到类似的破坏。我们还在无细胞人类 DNA 数据中鉴定出诊断性亚核小体颗粒残留物,作为凋亡细胞转录基因的遗迹。因此,从核酸酶保护数据中鉴定亚核小体片段代表了结构表观基因组学的一般策略。











































 京公网安备 11010802027423号
京公网安备 11010802027423号