当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Binding Mechanisms of Electron Transport Proteins with Cyanobacterial Photosystem I: An Integrated Computational and Experimental Model
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-01-16 00:00:00 , DOI: 10.1021/acs.jpcb.7b08307 Karan Kapoor 1, 2 , Derek J. Cashman 3 , Luke Nientimp 3 , Barry D. Bruce 1 , Jerome Baudry 1, 2
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-01-16 00:00:00 , DOI: 10.1021/acs.jpcb.7b08307 Karan Kapoor 1, 2 , Derek J. Cashman 3 , Luke Nientimp 3 , Barry D. Bruce 1 , Jerome Baudry 1, 2
Affiliation
|
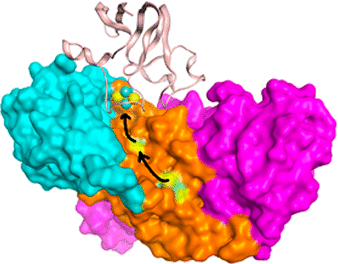
The stromal domain (PsaC, D, and E) of photosystem I (PSI) in cyanobacteria accepts electrons from PsaA and PsaB of photosystem I (PSI). These electrons are then used in the reduction of transiently bound ferredoxin (Fd) or flavodoxin. Experimental X-ray and NMR structures are known for all of these protein partners separately, yet to date, there is no known experimental structure of the PSI/Fd complexes in the published literature. Computational models of Fd docked with the stromal domain of cyanobacterial PSI were assembled here starting from X-ray and NMR structures of PSI and Fd. Predicted models of specific regions of protein–protein interactions were built on the basis of energetic frustration, residue conservation and evolutionary importance, as well as from experimental site-directed mutagenesis and cross-linking studies. Microsecond time-scale molecular dynamics simulations of the PSI/Fd complexes suggest, rather than a single complex structure, the possible existence of multiple transient complexes of Fd bound to PSI.
中文翻译:

电子转运蛋白与蓝细菌光系统的结合机理I:集成的计算和实验模型
蓝细菌中光系统I(PSI)的基质结构域(PsaC,D和E)接受来自光系统I(PSI)的PsaA和PsaB的电子。然后将这些电子用于还原瞬时结合的铁氧还蛋白(Fd)或黄酮毒素。所有这些蛋白伴侣的实验X射线和NMR结构都是已知的,但迄今为止,在公开的文献中还没有已知的PSI / Fd络合物的实验结构。在此,从PSI和Fd的X射线和NMR结构出发,组装了与蓝藻PSI的基质结构域对接的Fd的计算模型。蛋白质-蛋白质相互作用特定区域的预测模型是基于能量挫败,残基保守性和进化重要性以及实验定点诱变和交联研究建立的。
更新日期:2018-01-16
中文翻译:

电子转运蛋白与蓝细菌光系统的结合机理I:集成的计算和实验模型
蓝细菌中光系统I(PSI)的基质结构域(PsaC,D和E)接受来自光系统I(PSI)的PsaA和PsaB的电子。然后将这些电子用于还原瞬时结合的铁氧还蛋白(Fd)或黄酮毒素。所有这些蛋白伴侣的实验X射线和NMR结构都是已知的,但迄今为止,在公开的文献中还没有已知的PSI / Fd络合物的实验结构。在此,从PSI和Fd的X射线和NMR结构出发,组装了与蓝藻PSI的基质结构域对接的Fd的计算模型。蛋白质-蛋白质相互作用特定区域的预测模型是基于能量挫败,残基保守性和进化重要性以及实验定点诱变和交联研究建立的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号