当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reverse mapping method for complex polymer systems
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-12-06 , DOI: 10.1002/jcc.25129 Jakub Krajniak 1 , Zidan Zhang 2 , Sudharsan Pandiyan 2 , Eric Nies 2 , Giovanni Samaey 1
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-12-06 , DOI: 10.1002/jcc.25129 Jakub Krajniak 1 , Zidan Zhang 2 , Sudharsan Pandiyan 2 , Eric Nies 2 , Giovanni Samaey 1
Affiliation
|
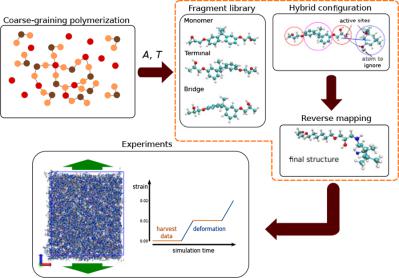
We present a comprehensive approach for reverse mapping of complex polymer systems in which the connectivity is created by the simulation of chemical reactions at the coarse‐grained scale. Within the work, we use a recently developed generic adaptive reverse mapping procedure that we adapt to handle the varying connectivity structure resulting from the chemical reactions. The method is independent of the coarse‐grained and fine‐grained force‐fields by design and relies only on a single control parameter. We employ the approach to reverse map four different systems: a three‐component epoxy network, a trimethylol melamine network, a hyperbranched polymer, and a polyethylene terephthalate. In the case of the epoxy network, we use two fine‐scale representations: fully atomistic and united‐atom. Whereas the trimethylol melamine network the hyperbranched polymer and the polyethylene terephthalate are reverse mapped to the fully atomistic description. After the reverse mapping, we examine the fine‐grained structure by comparing the radial distribution functions with respect to the control parameter. Moreover, in the case of the epoxy we perform tensile‐test experiments and examine the resulting Young's modulus. In all cases, we show how the properties of the reverse mapped systems depend on the control parameters. In general, we see that the results are relatively insensitive to the control parameter and the resulting atomistic systems are stable. Only for the trimethylol melamine network, we notice chemically incorrect conformations when the reverse mapping is performed too fast. We provide a remedy for this issue. © 2017 Wiley Periodicals, Inc.
中文翻译:

复杂聚合物体系的逆向映射方法
我们提出了一种复杂聚合物系统反向映射的综合方法,其中连接性是通过在粗粒度尺度上模拟化学反应来创建的。在这项工作中,我们使用了最近开发的通用自适应反向映射程序,我们可以适应处理由化学反应引起的不同连接结构。该方法在设计上独立于粗粒度和细粒度力场,仅依赖于单个控制参数。我们采用这种方法来反向映射四种不同的系统:三组分环氧树脂网络、三羟甲基三聚氰胺网络、超支化聚合物和聚对苯二甲酸乙二醇酯。在环氧树脂网络的情况下,我们使用两种精细表示:完全原子和联合原子。而三羟甲基三聚氰胺网络,超支化聚合物和聚对苯二甲酸乙二醇酯被反向映射到完全原子描述。在反向映射之后,我们通过比较径向分布函数与控制参数来检查细粒度结构。此外,在环氧树脂的情况下,我们进行拉伸测试实验并检查由此产生的杨氏模量。在所有情况下,我们展示了反向映射系统的属性如何取决于控制参数。一般来说,我们看到结果对控制参数相对不敏感,并且由此产生的原子系统是稳定的。仅对于三羟甲基三聚氰胺网络,当反向映射执行得太快时,我们注意到化学上不正确的构象。我们为此问题提供了补救措施。
更新日期:2017-12-06
中文翻译:

复杂聚合物体系的逆向映射方法
我们提出了一种复杂聚合物系统反向映射的综合方法,其中连接性是通过在粗粒度尺度上模拟化学反应来创建的。在这项工作中,我们使用了最近开发的通用自适应反向映射程序,我们可以适应处理由化学反应引起的不同连接结构。该方法在设计上独立于粗粒度和细粒度力场,仅依赖于单个控制参数。我们采用这种方法来反向映射四种不同的系统:三组分环氧树脂网络、三羟甲基三聚氰胺网络、超支化聚合物和聚对苯二甲酸乙二醇酯。在环氧树脂网络的情况下,我们使用两种精细表示:完全原子和联合原子。而三羟甲基三聚氰胺网络,超支化聚合物和聚对苯二甲酸乙二醇酯被反向映射到完全原子描述。在反向映射之后,我们通过比较径向分布函数与控制参数来检查细粒度结构。此外,在环氧树脂的情况下,我们进行拉伸测试实验并检查由此产生的杨氏模量。在所有情况下,我们展示了反向映射系统的属性如何取决于控制参数。一般来说,我们看到结果对控制参数相对不敏感,并且由此产生的原子系统是稳定的。仅对于三羟甲基三聚氰胺网络,当反向映射执行得太快时,我们注意到化学上不正确的构象。我们为此问题提供了补救措施。









































 京公网安备 11010802027423号
京公网安备 11010802027423号