当前位置:
X-MOL 学术
›
ChemMedChem
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
AquaMMapS: An Alternative Tool to Monitor the Role of Water Molecules During Protein–Ligand Association
ChemMedChem ( IF 3.6 ) Pub Date : 2018-01-25 , DOI: 10.1002/cmdc.201700564 Alberto Cuzzolin 1 , Giuseppe Deganutti 1 , Veronica Salmaso 1 , Mattia Sturlese 1 , Stefano Moro 1
ChemMedChem ( IF 3.6 ) Pub Date : 2018-01-25 , DOI: 10.1002/cmdc.201700564 Alberto Cuzzolin 1 , Giuseppe Deganutti 1 , Veronica Salmaso 1 , Mattia Sturlese 1 , Stefano Moro 1
Affiliation
|
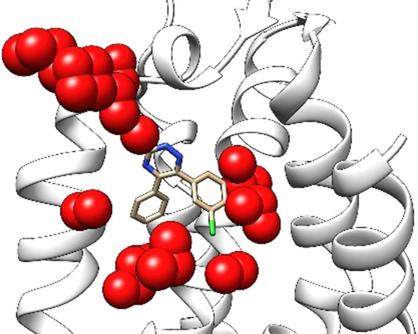
Unquestionably, water appears to be an active player in the noncovalent protein–ligand binding process, as it can either bridge interactions between protein and ligand or can be replaced by the bound ligand. Accordingly, in the last decade, alternative computational methodologies have been sought with the aim of predicting the position and thermodynamic profile of water molecules (i.e., hydration sites) in the binding site using either the ligand‐bound or ligand‐free protein conformation. Herein, we present an alternative approach, named AquaMMapS, that provides a three‐dimensional sampling of putative hydration sites. Interestingly, AquaMMapS can post‐inspect molecular dynamics (MD) trajectories obtained from different MD engines using indifferently crystallographic or docking‐driven structures as a starting point. Moreover, AquaMMapS is naturally integrated into supervised molecular dynamics (SuMD) simulations, presenting the possibility to inspect hydration sites during the ligand–protein association process. Finally, a penalty scoring method, named AquaMMapScoring(AMS), was developed to evaluate the number and nature of the water molecules displaced by a ligand approaching its binding site during the binding event, guiding a medicinal chemist to explore the most suitable regions of a ligand that can be decorated either with or without interfering with the interaction networks mediated by water molecules with specific recognition regions of the protein.
中文翻译:

AquaMMapS:在蛋白质-配体缔合过程中监测水分子作用的替代工具
毫无疑问,水似乎是非共价蛋白-配体结合过程的积极参与者,因为它可以桥接蛋白和配体之间的相互作用,也可以被结合的配体代替。因此,在过去的十年中,人们一直在寻找其他计算方法,目的是使用结合有配体或无配体的蛋白质构象预测结合位点中水分子(即水合位点)的位置和热力学特征。在此,我们提出了另一种方法,称为AquaMMapS,它提供了假定水化位点的三维采样。有趣的是,AquaMMapS可以使用不同的晶体学或对接驱动的结构作为起点,对从不同MD引擎获得的分子动力学(MD)轨迹进行后检查。而且,AquaMMapS自然地集成到有监督的分子动力学(SuMD)模拟中,从而提供了在配体与蛋白质缔合过程中检查水合位点的可能性。最后,开发了一种罚分方法,称为AquaMMapScoring(AMS),以评估在结合过程中被配体接近其结合位点的水分子置换的水分子的数量和性质,指导药物化学家探索最合适的区域。可以修饰的配体,可以干扰或不干扰由水分子与蛋白质的特定识别区域介导的相互作用网络。
更新日期:2018-01-25
中文翻译:

AquaMMapS:在蛋白质-配体缔合过程中监测水分子作用的替代工具
毫无疑问,水似乎是非共价蛋白-配体结合过程的积极参与者,因为它可以桥接蛋白和配体之间的相互作用,也可以被结合的配体代替。因此,在过去的十年中,人们一直在寻找其他计算方法,目的是使用结合有配体或无配体的蛋白质构象预测结合位点中水分子(即水合位点)的位置和热力学特征。在此,我们提出了另一种方法,称为AquaMMapS,它提供了假定水化位点的三维采样。有趣的是,AquaMMapS可以使用不同的晶体学或对接驱动的结构作为起点,对从不同MD引擎获得的分子动力学(MD)轨迹进行后检查。而且,AquaMMapS自然地集成到有监督的分子动力学(SuMD)模拟中,从而提供了在配体与蛋白质缔合过程中检查水合位点的可能性。最后,开发了一种罚分方法,称为AquaMMapScoring(AMS),以评估在结合过程中被配体接近其结合位点的水分子置换的水分子的数量和性质,指导药物化学家探索最合适的区域。可以修饰的配体,可以干扰或不干扰由水分子与蛋白质的特定识别区域介导的相互作用网络。











































 京公网安备 11010802027423号
京公网安备 11010802027423号