Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DNAzyme Based Nanomachine for in Situ Detection of MicroRNA in Living Cells
ACS Sensors ( IF 8.2 ) Pub Date : 2017-12-12 00:00:00 , DOI: 10.1021/acssensors.7b00710 Jing Liu 1, 2 , Meirong Cui 1, 3 , Hong Zhou 1, 2 , Wenrong Yang 1, 2
ACS Sensors ( IF 8.2 ) Pub Date : 2017-12-12 00:00:00 , DOI: 10.1021/acssensors.7b00710 Jing Liu 1, 2 , Meirong Cui 1, 3 , Hong Zhou 1, 2 , Wenrong Yang 1, 2
Affiliation
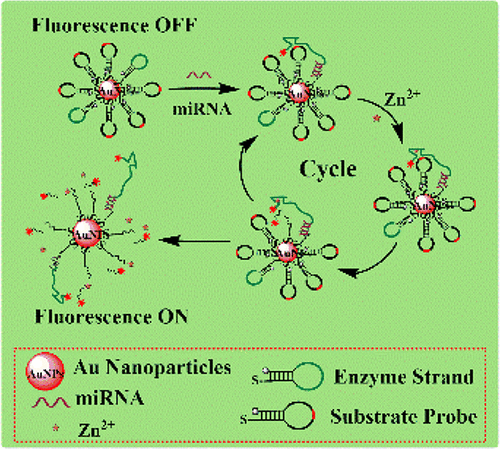
|
The capability of in situ detection of microRNA in living cells with signal amplification strategy is of fundamental importance, and it will open up a new opportunity in development of diagnosis and prognosis of many diseases. Herein we report a swing DNA nanomachine for intracellular microRNA detection. The surfaces of Au nanoparticles (NPs) are modified by two hairpin DNA. We observe that one DNA (MB2) will open its hairpin structure upon partial hybridization with target miR-21 after entering into cells, and the other part of its hairpin structure could further react with the other hairpin DNA (MB1) to form a Zn2+-specific DNAzyme. This results in the disruption of MB1 through shearing action and the release of fluorescein Cy5. To provide an intelligent DNA nanomachine, MB2 is available again with the shearing action to bind with MB1, which provides effective signal amplification. This target-responsive, DNA nanomachine-based method showed a detection limit of 0.1 nM in vitro, and this approach could be an important step toward intracellular amplified detection and imaging of various analytes in living cells.
中文翻译:

基于DNAzyme的纳米机用于活细胞中MicroRNA的原位检测
利用信号放大策略在活细胞中原位检测微小RNA的能力至关重要,它将为发展许多疾病的诊断和预后开辟新的机会。在本文中,我们报告了一种用于细胞内microRNA检测的Swing DNA纳米机。金纳米颗粒(NPs)的表面被两个发夹DNA修饰。我们观察到一个DNA(MB2)在进入细胞后与靶标miR-21部分杂交后将打开其发夹结构,其另一部分的发夹结构可能进一步与其他发夹DNA(MB1)反应形成Zn 2 +特异的DNA核酶。这导致通过剪切作用破坏MB1和释放荧光素Cy5。为了提供智能的DNA纳米机器,MB2再次具有剪切作用,可以与MB1结合,从而提供有效的信号放大。这种基于靶标的,基于DNA纳米机的方法在体外的检测极限为0.1 nM ,这种方法可能是朝着细胞内扩增检测和活细胞中各种分析物成像迈出的重要一步。
更新日期:2017-12-12
中文翻译:

基于DNAzyme的纳米机用于活细胞中MicroRNA的原位检测
利用信号放大策略在活细胞中原位检测微小RNA的能力至关重要,它将为发展许多疾病的诊断和预后开辟新的机会。在本文中,我们报告了一种用于细胞内microRNA检测的Swing DNA纳米机。金纳米颗粒(NPs)的表面被两个发夹DNA修饰。我们观察到一个DNA(MB2)在进入细胞后与靶标miR-21部分杂交后将打开其发夹结构,其另一部分的发夹结构可能进一步与其他发夹DNA(MB1)反应形成Zn 2 +特异的DNA核酶。这导致通过剪切作用破坏MB1和释放荧光素Cy5。为了提供智能的DNA纳米机器,MB2再次具有剪切作用,可以与MB1结合,从而提供有效的信号放大。这种基于靶标的,基于DNA纳米机的方法在体外的检测极限为0.1 nM ,这种方法可能是朝着细胞内扩增检测和活细胞中各种分析物成像迈出的重要一步。











































 京公网安备 11010802027423号
京公网安备 11010802027423号