当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-27 00:00:00 , DOI: 10.1021/acs.jproteome.7b00595 Fadhl M. Alakwaa 1 , Kumardeep Chaudhary 1 , Lana X. Garmire 1, 2
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-11-27 00:00:00 , DOI: 10.1021/acs.jproteome.7b00595 Fadhl M. Alakwaa 1 , Kumardeep Chaudhary 1 , Lana X. Garmire 1, 2
Affiliation
|
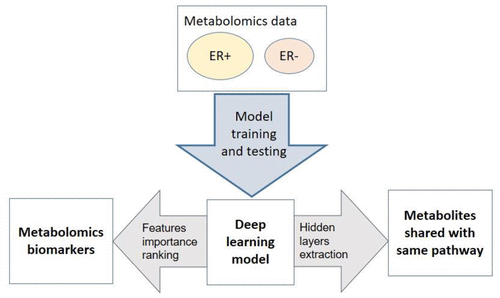
Metabolomics holds the promise as a new technology to diagnose highly heterogeneous diseases. Conventionally, metabolomics data analysis for diagnosis is done using various statistical and machine learning based classification methods. However, it remains unknown if deep neural network, a class of increasingly popular machine learning methods, is suitable to classify metabolomics data. Here we use a cohort of 271 breast cancer tissues, 204 positive estrogen receptor (ER+), and 67 negative estrogen receptor (ER−) to test the accuracies of feed-forward networks, a deep learning (DL) framework, as well as six widely used machine learning models, namely random forest (RF), support vector machines (SVM), recursive partitioning and regression trees (RPART), linear discriminant analysis (LDA), prediction analysis for microarrays (PAM), and generalized boosted models (GBM). DL framework has the highest area under the curve (AUC) of 0.93 in classifying ER+/ER– patients, compared to the other six machine learning algorithms. Furthermore, the biological interpretation of the first hidden layer reveals eight commonly enriched significant metabolomics pathways (adjusted P-value <0.05) that cannot be discovered by other machine learning methods. Among them, protein digestion and absorption and ATP-binding cassette (ABC) transporters pathways are also confirmed in integrated analysis between metabolomics and gene expression data in these samples. In summary, deep learning method shows advantages for metabolomics based breast cancer ER status classification, with both the highest prediction accuracy (AUC = 0.93) and better revelation of disease biology. We encourage the adoption of feed-forward networks based deep learning method in the metabolomics research community for classification.
中文翻译:

深度学习可准确预测乳腺癌代谢组学数据中的雌激素受体状态
代谢组学有望成为一种诊断高度异质性疾病的新技术。传统上,用于诊断的代谢组学数据分析是使用各种基于统计和机器学习的分类方法来完成的。然而,尚不知道深度神经网络(一类越来越流行的机器学习方法)是否适合对代谢组学数据进行分类。在这里,我们使用271个乳腺癌组织,204个雌激素阳性受体(ER +)和67个雌激素阴性受体(ER-)的队列来测试前馈网络,深度学习(DL)框架以及六个模型的准确性广泛使用的机器学习模型,即随机森林(RF),支持向量机(SVM),递归分区和回归树(RPART),线性判别分析(LDA),微阵列预测分析(PAM),和广义增强模型(GBM)。与其他六种机器学习算法相比,DL框架在对ER + / ER–患者进行分类时的曲线下面积(AUC)最高,为0.93。此外,对第一个隐藏层的生物学解释揭示了八个普遍丰富的重要代谢组学途径(已调整P值<0.05),是其他机器学习方法无法发现的。其中,在这些样品的代谢组学和基因表达数据之间的综合分析中,也证实了蛋白质的消化吸收和ATP结合盒(ABC)转运蛋白途径。总之,深度学习方法显示出基于代谢组学的乳腺癌ER状态分类的优势,具有最高的预测准确性(AUC = 0.93)和更好的疾病生物学启示。我们鼓励在代谢组学研究领域中采用基于前馈网络的深度学习方法进行分类。
更新日期:2017-11-28
中文翻译:

深度学习可准确预测乳腺癌代谢组学数据中的雌激素受体状态
代谢组学有望成为一种诊断高度异质性疾病的新技术。传统上,用于诊断的代谢组学数据分析是使用各种基于统计和机器学习的分类方法来完成的。然而,尚不知道深度神经网络(一类越来越流行的机器学习方法)是否适合对代谢组学数据进行分类。在这里,我们使用271个乳腺癌组织,204个雌激素阳性受体(ER +)和67个雌激素阴性受体(ER-)的队列来测试前馈网络,深度学习(DL)框架以及六个模型的准确性广泛使用的机器学习模型,即随机森林(RF),支持向量机(SVM),递归分区和回归树(RPART),线性判别分析(LDA),微阵列预测分析(PAM),和广义增强模型(GBM)。与其他六种机器学习算法相比,DL框架在对ER + / ER–患者进行分类时的曲线下面积(AUC)最高,为0.93。此外,对第一个隐藏层的生物学解释揭示了八个普遍丰富的重要代谢组学途径(已调整P值<0.05),是其他机器学习方法无法发现的。其中,在这些样品的代谢组学和基因表达数据之间的综合分析中,也证实了蛋白质的消化吸收和ATP结合盒(ABC)转运蛋白途径。总之,深度学习方法显示出基于代谢组学的乳腺癌ER状态分类的优势,具有最高的预测准确性(AUC = 0.93)和更好的疾病生物学启示。我们鼓励在代谢组学研究领域中采用基于前馈网络的深度学习方法进行分类。



























 京公网安备 11010802027423号
京公网安备 11010802027423号