Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identifying DNA mismatches at single-nucleotide resolution by probing individual surface potentials of DNA-capped nanoparticles
Nanoscale ( IF 5.8 ) Pub Date : 2017-11-06 00:00:00 , DOI: 10.1039/c7nr05250b Hyungbeen Lee 1, 2, 3, 4 , Sang Won Lee 4, 5, 6, 7 , Gyudo Lee 4, 5, 6, 7, 8 , Wonseok Lee 1, 2, 3, 4 , Kihwan Nam 4, 9, 10, 11 , Jeong Hoon Lee 4, 12, 13, 14 , Kyo Seon Hwang 4, 15, 16, 17, 18 , Jaemoon Yang 4, 19, 20, 21 , Hyeyoung Lee 2, 3, 4, 22 , Sangsig Kim 4, 6, 7, 12 , Sang Woo Lee 1, 2, 3, 4 , Dae Sung Yoon 4, 5, 6, 7
Nanoscale ( IF 5.8 ) Pub Date : 2017-11-06 00:00:00 , DOI: 10.1039/c7nr05250b Hyungbeen Lee 1, 2, 3, 4 , Sang Won Lee 4, 5, 6, 7 , Gyudo Lee 4, 5, 6, 7, 8 , Wonseok Lee 1, 2, 3, 4 , Kihwan Nam 4, 9, 10, 11 , Jeong Hoon Lee 4, 12, 13, 14 , Kyo Seon Hwang 4, 15, 16, 17, 18 , Jaemoon Yang 4, 19, 20, 21 , Hyeyoung Lee 2, 3, 4, 22 , Sangsig Kim 4, 6, 7, 12 , Sang Woo Lee 1, 2, 3, 4 , Dae Sung Yoon 4, 5, 6, 7
Affiliation
|
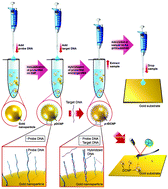
Here, we demonstrate a powerful method to discriminate DNA mismatches at single-nucleotide resolution from 0 to 5 mismatches (χ0 to χ5) using Kelvin probe force microscopy (KPFM). Using our previously developed method, we quantified the surface potentials (SPs) of individual DNA-capped nanoparticles (DCNPs, ∼100 nm). On each DCNP, DNA hybridization occurs between ∼2200 immobilized probe DNA (pDNA) and target DNA with mismatches (tDNA, ∼80 nM). Thus, each DCNP used in the bioassay (each pDNA–tDNA interaction) corresponds to a single ensemble in which a large number of pDNA–tDNA interactions take place. Moreover, one KPFM image can scan at least dozens of ensembles, which allows statistical analysis (i.e., an ensemble average) of many bioassay cases (ensembles) under the same conditions. We found that as the χn increased from χ0 to χ5 in the tDNA, the average SP of dozens of ensembles (DCNPs) was attenuated owing to fewer hybridization events between the pDNA and the tDNA. Remarkably, the SP attenuation vs. the χn showed an inverse-linear correlation, albeit the equilibrium constant for DNA hybridization exponentially decreased asymptotically as the χn increased. In addition, we observed a cascade reaction at a 100-fold lower concentration of tDNA (∼0.8 nM); the average SP of DCNPs exhibited no significant decrease but rather split into two separate states (no-hybridization vs. full-hybridization). Compared to complementary tDNA (i.e., χ0), the ratio of no-hybridization/full-hybridization within a given set of DCNPs became ∼1.6 times higher in the presence of tDNA with single mismatches (i.e., χ1). The results imply that our method opens new avenues not only in the research on the DNA hybridization mechanism in the presence of DNA mismatches but also in the development of a robust technology for DNA mismatch detection.
中文翻译:

通过探测DNA封端的纳米颗粒的单个表面电势来鉴定单核苷酸分辨率的DNA错配
这里,我们证明在单核苷酸分辨率从0至5个错配(一个功能强大的方法来判别DNA错配χ 0至χ 5),使用开尔文探针力显微镜(KPFM)。使用我们先前开发的方法,我们量化了单个DNA封端的纳米颗粒(DCNPs,〜100 nm)的表面电势(SP)。在每个DCNP上,DNA杂交发生在〜2200个固定的探针DNA(p DNA)和具有错配(t DNA,〜80 nM)的靶DNA之间。因此,每个DCNP在生物测定中使用(每个p DNA-吨DNA相互作用)对应于单个合奏在其中大量的p DNA-吨DNA相互作用发生。此外,一张KPFM图像可以扫描至少几十个集合,这允许在相同条件下对许多生物测定案例(集合)进行统计分析(即,集合平均值)。我们发现,当χ ñ从增加的χ 0至χ 5在T-DNA,几十合奏的平均SP(DCNPs)减弱由于较少之间的杂交事件p DNA并且吨DNA。值得注意的是,SP的衰减与该χ Ñ呈逆线性相关,尽管用于DNA杂交的平衡常数呈指数下降渐近作为χ ñ增加。此外,我们观察到级联反应在低100倍的t DNA浓度(约0.8 nM)下发生。DCNPs的平均SP没有显示出明显的下降,而是分为两个独立的状态(无杂交与完全杂交)。相比互补吨DNA(即,χ 0),一组给定的DCNPs内没有杂交/全杂交的比率在T-DNA的存在下,用单错配(成为~1.6倍即,χ 1)。结果表明,我们的方法不仅为存在DNA错配的DNA杂交机制研究开辟了新途径,而且为DNA错配检测的可靠技术的发展开辟了新的途径。
更新日期:2017-11-23
中文翻译:

通过探测DNA封端的纳米颗粒的单个表面电势来鉴定单核苷酸分辨率的DNA错配
这里,我们证明在单核苷酸分辨率从0至5个错配(一个功能强大的方法来判别DNA错配χ 0至χ 5),使用开尔文探针力显微镜(KPFM)。使用我们先前开发的方法,我们量化了单个DNA封端的纳米颗粒(DCNPs,〜100 nm)的表面电势(SP)。在每个DCNP上,DNA杂交发生在〜2200个固定的探针DNA(p DNA)和具有错配(t DNA,〜80 nM)的靶DNA之间。因此,每个DCNP在生物测定中使用(每个p DNA-吨DNA相互作用)对应于单个合奏在其中大量的p DNA-吨DNA相互作用发生。此外,一张KPFM图像可以扫描至少几十个集合,这允许在相同条件下对许多生物测定案例(集合)进行统计分析(即,集合平均值)。我们发现,当χ ñ从增加的χ 0至χ 5在T-DNA,几十合奏的平均SP(DCNPs)减弱由于较少之间的杂交事件p DNA并且吨DNA。值得注意的是,SP的衰减与该χ Ñ呈逆线性相关,尽管用于DNA杂交的平衡常数呈指数下降渐近作为χ ñ增加。此外,我们观察到级联反应在低100倍的t DNA浓度(约0.8 nM)下发生。DCNPs的平均SP没有显示出明显的下降,而是分为两个独立的状态(无杂交与完全杂交)。相比互补吨DNA(即,χ 0),一组给定的DCNPs内没有杂交/全杂交的比率在T-DNA的存在下,用单错配(成为~1.6倍即,χ 1)。结果表明,我们的方法不仅为存在DNA错配的DNA杂交机制研究开辟了新途径,而且为DNA错配检测的可靠技术的发展开辟了新的途径。











































 京公网安备 11010802027423号
京公网安备 11010802027423号