当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
B -factor profile prediction for RNA flexibility using support vector machines
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-11-21 , DOI: 10.1002/jcc.25124 Ivantha Guruge 1 , Ghazaleh Taherzadeh 1 , Jian Zhan 1 , Yaoqi Zhou 1 , Yuedong Yang 1, 2
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2017-11-21 , DOI: 10.1002/jcc.25124 Ivantha Guruge 1 , Ghazaleh Taherzadeh 1 , Jian Zhan 1 , Yaoqi Zhou 1 , Yuedong Yang 1, 2
Affiliation
|
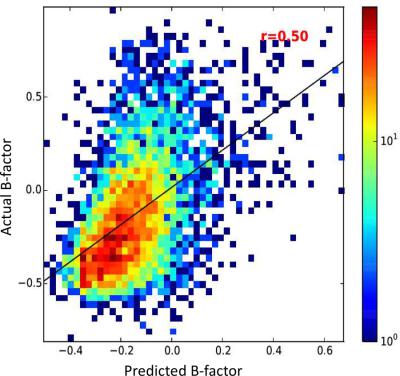
Determining the flexibility of structured biomolecules is important for understanding their biological functions. One quantitative measurement of flexibility is the atomic Debye‐Waller factor or temperature B‐factor. Most existing studies are limited to temperature B‐factors of proteins and their prediction. Only one method attempted to predict temperature B‐factors of ribosomal RNA. Here, we developed and compared machine‐learning techniques in prediction of temperature B‐factors of RNAs. The best model based on Support Vector Machines yields Pearson's correction coefficient at 0.51 for fivefold cross validation and 0.50 for the independent test. Analysis of the performance indicates that the model has the best performance on rRNAs, tRNAs, and protein‐bound RNAs, for long chains in particular. The server is available at http://sparks-lab.org/server/RNAflex. © 2017 Wiley Periodicals, Inc.
中文翻译:

使用支持向量机对 RNA 灵活性进行 B 因子分布预测
确定结构化生物分子的灵活性对于理解它们的生物学功能很重要。柔韧性的一种定量测量是原子德拜-沃勒因子或温度 B 因子。大多数现有研究仅限于蛋白质的温度 B 因子及其预测。只有一种方法试图预测核糖体 RNA 的温度 B 因子。在这里,我们开发并比较了预测 RNA 温度 B 因素的机器学习技术。基于支持向量机的最佳模型产生的 Pearson 校正系数对于五重交叉验证为 0.51,对于独立测试为 0.50。性能分析表明,该模型在 rRNA、tRNA 和蛋白质结合的 RNA 上具有最佳性能,尤其是长链。该服务器可从 http://sparks-lab 获得。组织/服务器/RNAflex。© 2017 威利期刊公司。
更新日期:2017-11-21
中文翻译:

使用支持向量机对 RNA 灵活性进行 B 因子分布预测
确定结构化生物分子的灵活性对于理解它们的生物学功能很重要。柔韧性的一种定量测量是原子德拜-沃勒因子或温度 B 因子。大多数现有研究仅限于蛋白质的温度 B 因子及其预测。只有一种方法试图预测核糖体 RNA 的温度 B 因子。在这里,我们开发并比较了预测 RNA 温度 B 因素的机器学习技术。基于支持向量机的最佳模型产生的 Pearson 校正系数对于五重交叉验证为 0.51,对于独立测试为 0.50。性能分析表明,该模型在 rRNA、tRNA 和蛋白质结合的 RNA 上具有最佳性能,尤其是长链。该服务器可从 http://sparks-lab 获得。组织/服务器/RNAflex。© 2017 威利期刊公司。











































 京公网安备 11010802027423号
京公网安备 11010802027423号