当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mutation-Induced Deamidation of Corneal Dystrophy-Related Transforming Growth Factor β-Induced Protein
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-22 00:00:00 , DOI: 10.1021/acs.biochem.7b00668 Nadia Sukusu Nielsen 1 , Dennis Wilkens Juhl 1 , Ebbe Toftgaard Poulsen 1 , Marie V. Lukassen 1 , Emil Christian Poulsen 1 , Michael W. Risør 1 , Carsten Scavenius 1 , Jan J. Enghild 1
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-22 00:00:00 , DOI: 10.1021/acs.biochem.7b00668 Nadia Sukusu Nielsen 1 , Dennis Wilkens Juhl 1 , Ebbe Toftgaard Poulsen 1 , Marie V. Lukassen 1 , Emil Christian Poulsen 1 , Michael W. Risør 1 , Carsten Scavenius 1 , Jan J. Enghild 1
Affiliation
|
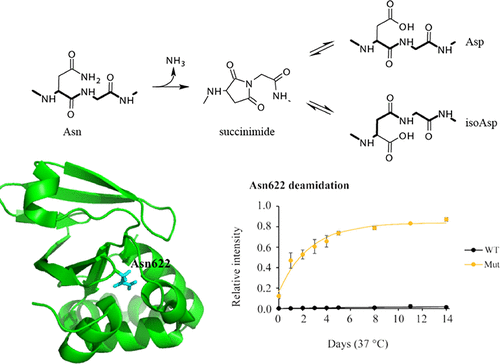
Mutations in the transforming growth factor β-induced protein (TGFBIp) cause phenotypically diverse corneal dystrophies, where protein aggregation in the cornea leads to severe visual impairment. Previous studies have shown a relationship between mutant-specific corneal dystrophy phenotypes and the thermodynamic stability of TGFBIp. Using liquid chromatography–tandem mass spectrometry and nuclear magnetic resonance (NMR), we investigated correlations between the structural integrity of disease-related mutants of the fourth FAS1 domain (FAS1-4) and deamidation of TGFBIp residue Asn622. We observed a high rate of Asn622 deamidation in the A546D and A546D/P551Q FAS1-4 mutants that were both largely unstructured as determined by NMR. Conversely, the more structurally organized A546T and V624M FAS1-4 mutants had reduced deamidation rates, suggesting that a folded and stable FAS1-4 domain precludes Asn622 deamidation. Wild-type, R555Q, and R555W FAS1-4 mutants displayed very slow deamidation, which agrees with their similar and ordered NMR structures, where Asn622 is in a locked conformation. We confirmed the FAS1-4 mutational effect on deamidation rates in full-length TGFBIp mutants and found a similar ranking compared to that of the FAS1-4 domain alone. Consequently, the deamidation rate of Asn622 can be used to predict the structural effect of the many destabilizing and/or stabilizing mutations reported for TGFBIp. In addition, the deamidation of Asn622 may influence the pathophysiology of TGFBIp-induced corneal dystrophies.
中文翻译:

突变诱导的角膜营养不良相关转化生长因子β诱导蛋白脱酰胺
转化生长因子β诱导蛋白(TGFBIp)的突变会导致表型多样化的角膜营养不良,其中角膜中的蛋白质聚集会导致严重的视力障碍。先前的研究表明突变体特异性角膜营养不良表型与TGFBIp的热力学稳定性之间的关系。使用液相色谱-串联质谱和核磁共振(NMR),我们研究了与疾病相关的第四个FAS1域(FAS1-4)突变体的结构完整性与TGFBIp残基Asn622脱酰胺化之间的相关性。我们观察到在A546D和A546D / P551Q FAS1-4突变体中,Asn622的酰胺化率很高,这两个突变体都是通过NMR测得的大部分非结构化结构。相反,结构更为组织化的A546T和V624M FAS1-4突变体的脱酰胺作用降低,提示折叠和稳定的FAS1-4结构域可防止Asn622脱酰胺。野生型,R555Q和R555W FAS1-4突变体显示出非常缓慢的脱酰胺作用,这与它们的相似和有序NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。这与它们相似且有序的NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。这与它们相似且有序的NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。Asn622的脱酰胺率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。Asn622的脱酰胺率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。
更新日期:2017-11-22
中文翻译:

突变诱导的角膜营养不良相关转化生长因子β诱导蛋白脱酰胺
转化生长因子β诱导蛋白(TGFBIp)的突变会导致表型多样化的角膜营养不良,其中角膜中的蛋白质聚集会导致严重的视力障碍。先前的研究表明突变体特异性角膜营养不良表型与TGFBIp的热力学稳定性之间的关系。使用液相色谱-串联质谱和核磁共振(NMR),我们研究了与疾病相关的第四个FAS1域(FAS1-4)突变体的结构完整性与TGFBIp残基Asn622脱酰胺化之间的相关性。我们观察到在A546D和A546D / P551Q FAS1-4突变体中,Asn622的酰胺化率很高,这两个突变体都是通过NMR测得的大部分非结构化结构。相反,结构更为组织化的A546T和V624M FAS1-4突变体的脱酰胺作用降低,提示折叠和稳定的FAS1-4结构域可防止Asn622脱酰胺。野生型,R555Q和R555W FAS1-4突变体显示出非常缓慢的脱酰胺作用,这与它们的相似和有序NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。这与它们相似且有序的NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。这与它们相似且有序的NMR结构一致,其中Asn622处于锁定构象。我们证实了FAS1-4突变对全长TGFBIp突变体中脱酰胺作用的影响,并且与单独的FAS1-4结构域相比,发现了相似的排名。因此,Asn622的脱酰胺速率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。Asn622的脱酰胺率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。Asn622的脱酰胺率可用于预测TGFBIp报道的许多不稳定和/或稳定突变的结构效应。此外,Asn622的脱酰胺作用可能会影响TGFBIp诱导的角膜营养不良的病理生理。









































 京公网安备 11010802027423号
京公网安备 11010802027423号