当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Quantum Mechanics/Molecular Mechanics Simulations Identify the Ring-Opening Mechanism of Creatininase
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.biochem.7b01032 Jitrayut Jitonnom 1 , Jon I. Mujika 2 , Marc W. van der Kamp 3, 4 , Adrian J. Mulholland 4
Biochemistry ( IF 2.9 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.biochem.7b01032 Jitrayut Jitonnom 1 , Jon I. Mujika 2 , Marc W. van der Kamp 3, 4 , Adrian J. Mulholland 4
Affiliation
|
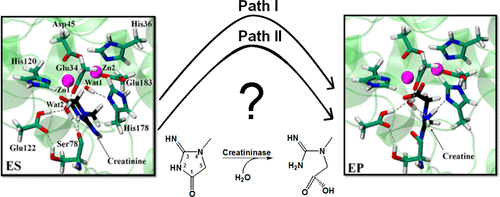
Creatininase catalyzes the conversion of creatinine (a biosensor for kidney function) to creatine via a two-step mechanism: water addition followed by ring opening. Water addition is common to other known cyclic amidohydrolases, but the precise mechanism for ring opening is still under debate. The proton donor in this step is either His178 or a water molecule bound to one of the metal ions, and the roles of His178 and Glu122 are unclear. Here, the two possible reaction pathways have been fully examined by means of combined quantum mechanics/molecular mechanics simulations at the SCC-DFTB/CHARMM22 level of theory. The results indicate that His178 is the main catalytic residue for the whole reaction and explain its role as proton shuttle during the ring-opening step. In the first step, His178 provides electrostatic stabilization to the gem-diolate tetrahedral intermediate. In the second step, His178 abstracts the hydroxyl proton of the intermediate and delivers it to the cyclic amide nitrogen, leading to ring opening. The latter is the rate-limiting step with a free energy barrier of 18.5 kcal/mol, in agreement with the experiment. We find that Glu122 must be protonated during the enzyme reaction, so that it can form a stable hydrogen bond with its neighboring water molecule. Simulations of the E122Q mutant showed that this replacement disrupts the H-bond network formed by three conserved residues (Glu34, Ser78, and Glu122) and water, increasing the energy barrier. Our computational studies provide a comprehensive explanation for previous structural and kinetic observations, including why the H178A mutation causes a complete loss of activity but the E122Q mutation does not.
中文翻译:

量子力学/分子力学模拟确定肌酐酶的开环机理
肌酐酶通过两步机制催化肌酸酐(一种肾功能的生物传感器)向肌酸的转化:加水然后开环。加水是其他已知的环状酰胺水解酶所共有的,但是开环的确切机理仍在争论中。在此步骤中,质子供体是His178或与一种金属离子结合的水分子,His178和Glu122的作用尚不清楚。在这里,通过在SCC-DFTB / CHARMM22理论水平上的组合量子力学/分子力学模拟,已经充分检查了两种可能的反应途径。结果表明,His178是整个反应的主要催化残基,并解释了其在开环步骤中作为质子穿梭的作用。第一步,His178为羟甲酸酯四面体中间体。在第二步中,His178提取中间体的羟基质子,并将其传递至环状酰胺氮,导致开环。与实验一致,后者是具有18.5 kcal / mol的自由能垒的限速步骤。我们发现,Glu122必须在酶反应过程中被质子化,以便它可以与其相邻的水分子形成稳定的氢键。对E122Q突变体的模拟表明,这种置换破坏了由三个保守残基(Glu34,Ser78和Glu122)和水形成的H键网络,从而增加了能量垒。我们的计算研究为以前的结构和动力学观察提供了全面的解释,包括为什么H178A突变会导致活性完全丧失,而E122Q突变却不会。
更新日期:2017-11-22
中文翻译:

量子力学/分子力学模拟确定肌酐酶的开环机理
肌酐酶通过两步机制催化肌酸酐(一种肾功能的生物传感器)向肌酸的转化:加水然后开环。加水是其他已知的环状酰胺水解酶所共有的,但是开环的确切机理仍在争论中。在此步骤中,质子供体是His178或与一种金属离子结合的水分子,His178和Glu122的作用尚不清楚。在这里,通过在SCC-DFTB / CHARMM22理论水平上的组合量子力学/分子力学模拟,已经充分检查了两种可能的反应途径。结果表明,His178是整个反应的主要催化残基,并解释了其在开环步骤中作为质子穿梭的作用。第一步,His178为羟甲酸酯四面体中间体。在第二步中,His178提取中间体的羟基质子,并将其传递至环状酰胺氮,导致开环。与实验一致,后者是具有18.5 kcal / mol的自由能垒的限速步骤。我们发现,Glu122必须在酶反应过程中被质子化,以便它可以与其相邻的水分子形成稳定的氢键。对E122Q突变体的模拟表明,这种置换破坏了由三个保守残基(Glu34,Ser78和Glu122)和水形成的H键网络,从而增加了能量垒。我们的计算研究为以前的结构和动力学观察提供了全面的解释,包括为什么H178A突变会导致活性完全丧失,而E122Q突变却不会。











































 京公网安备 11010802027423号
京公网安备 11010802027423号