当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Patterns of Ligands Coordinated to Metallocofactors Extracted from the Protein Data Bank
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jcim.7b00468 Luca Belmonte 1 , Sheref S. Mansy 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2017-11-21 00:00:00 , DOI: 10.1021/acs.jcim.7b00468 Luca Belmonte 1 , Sheref S. Mansy 1
Affiliation
|
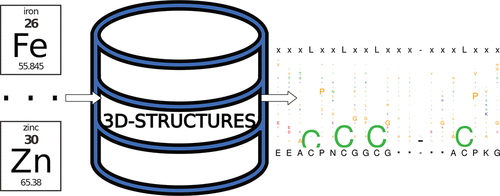
A new R tool is described that rapidly identifies, ranks, and clusters sequence patterns coordinated to metallocofactors. This tool, PdPDB, fills a void because, unlike currently available tools, PdPDB searches through sequences with metal coordination as the primary determinant and can identify patterns consisting of amino acids, nucleotides, and small molecule ligands at once. PdPDB was tested by analyzing structures that coordinate Fe2+/3+, [2Fe-2S], [4Fe-4S], Zn2+, and Mg2+ cofactors. PdPDB confirmed previously identified sequence motifs and revealed which residues are enriched (e.g., glycine) and are under-represented (e.g., glutamine) near ligands to metal centers. The data show the similarities and differences between different metal-binding sites. The patterns that coordinate metallocofactors vary, depending upon whether the metal ions play a structural or catalytic role, with catalytic metal centers exhibiting partial coordination by small molecule ligands. PdPDB 2.0.1 is freely available as a CRAN package.
中文翻译:

从蛋白质数据库中提取的与金属因子配体的配体模式
描述了一种新的R工具,该工具可以快速识别,排序和聚类与金属因子配合的序列模式。PdPDB这个工具填补了空白,因为与当前可用的工具不同,PdPDB搜索以金属配位作为主要决定因素的序列,并且可以一次识别出由氨基酸,核苷酸和小分子配体组成的模式。通过分析协调Fe 2 + / 3 +,[2Fe-2S],[4Fe-4S],Zn 2+和Mg 2+的结构来测试PdPDB辅助因子。PdPDB证实了先前确定的序列基序,并揭示了在金属中心配体附近哪些残基富集(例如甘氨酸)而代表性不足(例如谷氨酰胺)。数据显示了不同金属结合位点之间的异同。取决于金属离子起结构或催化作用,配位金属因子的模式变化,其中催化金属中心通过小分子配体表现出部分配位。PdPDB 2.0.1可作为CRAN软件包免费提供。
更新日期:2017-11-22
中文翻译:

从蛋白质数据库中提取的与金属因子配体的配体模式
描述了一种新的R工具,该工具可以快速识别,排序和聚类与金属因子配合的序列模式。PdPDB这个工具填补了空白,因为与当前可用的工具不同,PdPDB搜索以金属配位作为主要决定因素的序列,并且可以一次识别出由氨基酸,核苷酸和小分子配体组成的模式。通过分析协调Fe 2 + / 3 +,[2Fe-2S],[4Fe-4S],Zn 2+和Mg 2+的结构来测试PdPDB辅助因子。PdPDB证实了先前确定的序列基序,并揭示了在金属中心配体附近哪些残基富集(例如甘氨酸)而代表性不足(例如谷氨酰胺)。数据显示了不同金属结合位点之间的异同。取决于金属离子起结构或催化作用,配位金属因子的模式变化,其中催化金属中心通过小分子配体表现出部分配位。PdPDB 2.0.1可作为CRAN软件包免费提供。











































 京公网安备 11010802027423号
京公网安备 11010802027423号