当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of Conserved Water Sites in Protein Structures for Drug Design
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2017-12-04 00:00:00 , DOI: 10.1021/acs.jcim.7b00443 Marko Jukič 1 , Janez Konc 2, 3 , Stanislav Gobec 1 , Dušanka Janežič 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2017-12-04 00:00:00 , DOI: 10.1021/acs.jcim.7b00443 Marko Jukič 1 , Janez Konc 2, 3 , Stanislav Gobec 1 , Dušanka Janežič 3
Affiliation
|
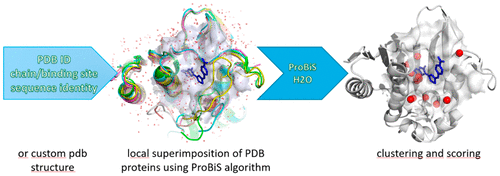
Identification of conserved waters in protein structures is a challenging task with applications in molecular docking and protein stability prediction. As an alternative to computationally demanding simulations of proteins in water, experimental cocrystallized waters in the Protein Data Bank (PDB) in combination with a local structure alignment algorithm can be used for reliable prediction of conserved water sites. We developed the ProBiS H2O approach based on the previously developed ProBiS algorithm, which enables identification of conserved water sites in proteins using experimental protein structures from the PDB or a set of custom protein structures available to the user. With a protein structure, a binding site, or an individual water molecule as a query, ProBiS H2O collects similar proteins from the PDB and performs local or binding site-specific superimpositions of the query structure with similar proteins using the ProBiS algorithm. It collects the experimental water molecules from the similar proteins and transposes them to the query protein. Transposed waters are clustered by their mutual proximity, which enables identification of discrete sites in the query protein with high water conservation. ProBiS H2O is a robust and fast new approach that uses existing experimental structural data to identify conserved water sites on the interfaces of protein complexes, for example protein–small molecule interfaces, and elsewhere on the protein structures. It has been successfully validated in several reported proteins in which conserved water molecules were found to play an important role in ligand binding with applications in drug design.
中文翻译:

蛋白质设计中蛋白质结构中保守水位的鉴定
在分子对接和蛋白质稳定性预测中的应用,鉴定蛋白质结构中的保守水域是一项艰巨的任务。作为对蛋白质的计算要求较高的模拟的替代方法,蛋白质数据库(PDB)中的实验性共结晶水与局部结构比对算法的结合可用于可靠预测保守的水位。我们基于以前开发的ProBiS算法开发了ProBiS H2O方法,该方法可以使用PDB的实验蛋白质结构或用户可用的一组定制蛋白质结构来鉴定蛋白质中的保守水位。以蛋白质结构,结合位点或单个水分子作为查询,ProBiS H2O从PDB收集相似的蛋白质,并使用ProBiS算法与相似的蛋白质执行查询结构的局部或结合位点特异性叠加。它从相似的蛋白质中收集实验水分子,并将其转置为查询蛋白质。换位的水通过它们的相互邻近性聚集在一起,从而可以在查询蛋白中识别出离散位点,并具有很高的节水率。ProBiS H2O是一种强大而快速的新方法,它使用现有的实验结构数据来识别蛋白质复合物界面(例如蛋白质-小分子界面)以及蛋白质结构上其他地方的保守水位。
更新日期:2017-12-04
中文翻译:

蛋白质设计中蛋白质结构中保守水位的鉴定
在分子对接和蛋白质稳定性预测中的应用,鉴定蛋白质结构中的保守水域是一项艰巨的任务。作为对蛋白质的计算要求较高的模拟的替代方法,蛋白质数据库(PDB)中的实验性共结晶水与局部结构比对算法的结合可用于可靠预测保守的水位。我们基于以前开发的ProBiS算法开发了ProBiS H2O方法,该方法可以使用PDB的实验蛋白质结构或用户可用的一组定制蛋白质结构来鉴定蛋白质中的保守水位。以蛋白质结构,结合位点或单个水分子作为查询,ProBiS H2O从PDB收集相似的蛋白质,并使用ProBiS算法与相似的蛋白质执行查询结构的局部或结合位点特异性叠加。它从相似的蛋白质中收集实验水分子,并将其转置为查询蛋白质。换位的水通过它们的相互邻近性聚集在一起,从而可以在查询蛋白中识别出离散位点,并具有很高的节水率。ProBiS H2O是一种强大而快速的新方法,它使用现有的实验结构数据来识别蛋白质复合物界面(例如蛋白质-小分子界面)以及蛋白质结构上其他地方的保守水位。









































 京公网安备 11010802027423号
京公网安备 11010802027423号