PLOS Neglected Tropical Diseases ( IF 3.8 ) Pub Date : 2017-11-20 , DOI: 10.1371/journal.pntd.0006075 Annika Brinkmann , Koray Ergünay , Aleksandar Radonić , Zeliha Kocak Tufan , Cristina Domingo , Andreas Nitsche
|
Background
We describe the development and evaluation of a novel method for targeted amplification and Next Generation Sequencing (NGS)-based identification of viral hemorrhagic fever (VHF) agents and assess the feasibility of this approach in diagnostics.
Methodology
An ultrahigh-multiplex panel was designed with primers to amplify all known variants of VHF-associated viruses and relevant controls. The performance of the panel was evaluated via serially quantified nucleic acids from Yellow fever virus, Rift Valley fever virus, Crimean-Congo hemorrhagic fever (CCHF) virus, Ebola virus, Junin virus and Chikungunya virus in a semiconductor-based sequencing platform. A comparison of direct NGS and targeted amplification-NGS was performed. The panel was further tested via a real-time nanopore sequencing-based platform, using clinical specimens from CCHF patients.
Principal findings
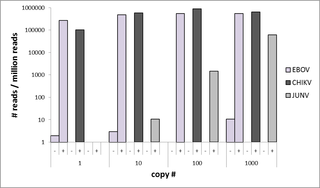
The multiplex primer panel comprises two pools of 285 and 256 primer pairs for the identification of 46 virus species causing hemorrhagic fevers, encompassing 6,130 genetic variants of the strains involved. In silico validation revealed that the panel detected over 97% of all known genetic variants of the targeted virus species. High levels of specificity and sensitivity were observed for the tested virus strains. Targeted amplification ensured viral read detection in specimens with the lowest virus concentration (1–10 genome equivalents) and enabled significant increases in specific reads over background for all viruses investigated. In clinical specimens, the panel enabled detection of the causative agent and its characterization within 10 minutes of sequencing, with sample-to-result time of less than 3.5 hours.
Conclusions
Virus enrichment via targeted amplification followed by NGS is an applicable strategy for the diagnosis of VHFs which can be adapted for high-throughput or nanopore sequencing platforms and employed for surveillance or outbreak monitoring.
中文翻译:

病毒性出血热诊断的多重扩增和下一代测序方法的开发和初步评估
背景
我们描述了一种新方法的开发和评估,该方法用于靶向扩增和基于下一代测序(NGS)的病毒性出血热(VHF)药物鉴定,并评估了该方法在诊断中的可行性。
方法
用引物设计超高多重面板,以扩增VHF相关病毒和相关对照的所有已知变体。在基于半导体的测序平台中,通过对黄热病病毒,裂谷热病毒,克里米亚-刚果出血热(CCHF)病毒,埃博拉病毒,朱宁病毒和基孔肯雅病毒的系列定量核酸进行评估,评估专家组的性能。进行了直接NGS和靶向扩增-NGS的比较。通过基于实时纳米孔测序的平台,使用来自CCHF患者的临床标本,对该面板进行了进一步测试。
主要发现
多重引物组包括两个由285和256个引物对组成的库,用于鉴定引起出血热的46种病毒,包括所涉菌株的6130种遗传变体。在计算机上进行的验证显示,专家组检测到了目标病毒物种所有已知遗传变异的97%以上。对于测试的病毒株,观察到高水平的特异性和敏感性。靶向扩增可确保在最低病毒浓度(1-10个基因组当量)的标本中检测病毒读数,并使所有研究病毒的特异性读数显着高于背景。在临床标本中,专家组能够在测序的10分钟内检测到病原体及其特征,样品到结果的时间不到3.5小时。
结论
通过靶向扩增和NGS富集病毒是诊断VHF的适用策略,可适用于高通量或纳米孔测序平台,并用于监视或暴发监测。



























 京公网安备 11010802027423号
京公网安备 11010802027423号