Bioorganic & Medicinal Chemistry Letters ( IF 2.5 ) Pub Date : 2017-11-04 , DOI: 10.1016/j.bmcl.2017.11.003 Zhen Wang , Li Ping Cheng , Xing Hua Zhang , Wan Pang , Liang Li , Jin Long Zhao
|
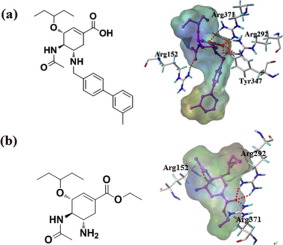
Neuraminidase (NA) is one of the particular potential targets for novel antiviral therapy. In this work, a series of neuraminidase inhibitors with the cyclohexene scaffold were studied based upon the combination of 3D-QSAR, molecular docking, and molecular dynamics techniques. The results indicate that the built 3D-QSAR models yield reliable statistical information: the correlation coefficient (r2) and cross-validation coefficient (q2) of CoMFA (comparative molecular field analysis) are 0.992 and 0.819; the r2 and q2 of CoMSIA (comparative molecular similarity analysis) are 0.992 and 0.863, respectively. Molecular docking and MD simulations were conducted to confirm the detailed binding mode of enzyme-inhibitor system. The new NA inhibitors had been designed, synthesized, and their inhibitory activities against group-1 neuraminidase were determined. One agent displayed excellent neuraminidase inhibition, with IC50 value of 39.6 μM against NA, while IC50 value for oseltamivir is 61.1 μM. This compound may be further investigated for the treatment of infection by the new type influenza virus.
中文翻译:

新的奥司他韦衍生物作为有效的神经氨酸酶抑制剂的设计,合成和生物学评估
神经氨酸酶(NA)是新型抗病毒治疗的特定潜在目标之一。在这项工作中,基于3D-QSAR,分子对接和分子动力学技术的结合,研究了一系列带有环己烯骨架的神经氨酸酶抑制剂。结果表明,所建立的3D-QSAR模型能够提供可靠的统计信息:CoMFA(比较分子场分析)的相关系数(r 2)和交叉验证系数(q 2)分别为0.992和0.819。r 2和q 2CoMSIA(比较分子相似性分析)的分别为0.992和0.863。进行了分子对接和MD模拟,以确认酶抑制剂系统的详细结合模式。设计,合成了新的NA抑制剂,并确定了它们对第1组神经氨酸酶的抑制活性。一种药剂表现出优异的神经氨酸酶抑制,IC 50 39.6μM针对NA的值,而IC 50为奥司他韦值是61.1μM。可以进一步研究该化合物用于治疗新型流感病毒的感染。











































 京公网安备 11010802027423号
京公网安备 11010802027423号