Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Characterizing the Motion of Jointed DNA Nanostructures Using a Coarse-Grained Model
ACS Nano ( IF 15.8 ) Pub Date : 2017-11-03 00:00:00 , DOI: 10.1021/acsnano.7b06470 Rahul Sharma 1 , John S. Schreck 2 , Flavio Romano 3 , Ard A. Louis 4 , Jonathan P. K. Doye 5
ACS Nano ( IF 15.8 ) Pub Date : 2017-11-03 00:00:00 , DOI: 10.1021/acsnano.7b06470 Rahul Sharma 1 , John S. Schreck 2 , Flavio Romano 3 , Ard A. Louis 4 , Jonathan P. K. Doye 5
Affiliation
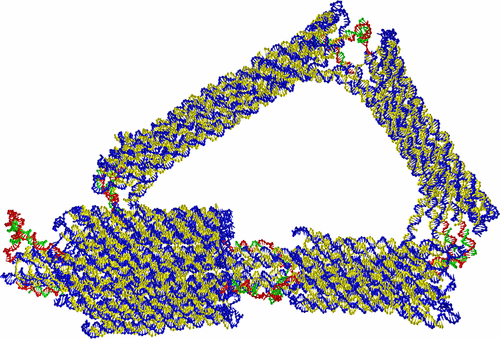
|
As detailed structural characterizations of large complex DNA nanostructures are hard to obtain experimentally, particularly if they have substantial flexibility, coarse-grained modeling can potentially provide an important complementary role. Such modeling can provide a detailed view of both the average structure and the structural fluctuations, as well as providing insight into how the nanostructure’s design determines its structural properties. Here, we present a case study of jointed DNA nanostructures using the oxDNA model. In particular, we consider archetypal hinge and sliding joints, as well as more complex structures involving a number of such coupled joints. Our results highlight how the nature of the motion in these structures can sensitively depend on the precise details of the joints. Furthermore, the generally good agreement with experiments illustrates the power of this approach and suggests the use of such modeling to prescreen the properties of putative designs.
中文翻译:

使用粗粒模型表征关节DNA纳米结构的运动
由于很难通过实验获得大型复杂DNA纳米结构的详细结构特征,尤其是如果它们具有很大的柔韧性时,粗粒度建模可能会提供重要的补充作用。这种建模可以提供平均结构和结构波动的详细视图,还可以洞悉纳米结构的设计如何确定其结构特性。在这里,我们介绍使用oxDNA模型的联合DNA纳米结构的案例研究。特别是,我们考虑了原型铰链和滑动接头,以及涉及许多此类联接接头的更复杂的结构。我们的结果强调了这些结构中运动的本质如何敏感地取决于关节的精确细节。此外,
更新日期:2017-11-05
中文翻译:

使用粗粒模型表征关节DNA纳米结构的运动
由于很难通过实验获得大型复杂DNA纳米结构的详细结构特征,尤其是如果它们具有很大的柔韧性时,粗粒度建模可能会提供重要的补充作用。这种建模可以提供平均结构和结构波动的详细视图,还可以洞悉纳米结构的设计如何确定其结构特性。在这里,我们介绍使用oxDNA模型的联合DNA纳米结构的案例研究。特别是,我们考虑了原型铰链和滑动接头,以及涉及许多此类联接接头的更复杂的结构。我们的结果强调了这些结构中运动的本质如何敏感地取决于关节的精确细节。此外,











































 京公网安备 11010802027423号
京公网安备 11010802027423号