当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Network-based modelling and percolation analysis of conformational dynamics and activation in the CDK2 and CDK4 proteins: dynamic and energetic polarization of the kinase lobes may determine divergence of the regulatory mechanisms
Molecular BioSystems Pub Date : 2017-09-12 00:00:00 , DOI: 10.1039/c7mb00355b G. M. Verkhivker 1, 2, 3, 4, 5
Molecular BioSystems Pub Date : 2017-09-12 00:00:00 , DOI: 10.1039/c7mb00355b G. M. Verkhivker 1, 2, 3, 4, 5
Affiliation
|
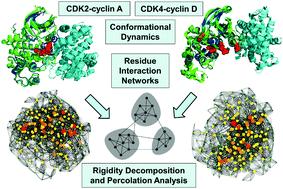
The overarching goal of delineating molecular principles underlying differentiation of the activation mechanisms in cyclin-dependent kinases (CDKs) is important for understanding regulatory divergences among closely related kinases which can be exploited in drug discovery of targeted and allosteric inhibitors. To systematically characterize dynamic, energetic and network signatures of the activation mechanisms, we combined atomistic simulations and elastic network modeling with the analysis of the residue interaction networks and rigidity decomposition of the CDK2-cyclin A and CDK4-cyclin D1/D3 complexes. The results of this study show that divergences in the activation mechanisms of CDK2 and CDK4 may be determined by differences in stabilization and allosteric cooperativity of the regulatory regions. We show that differential stabilization of the kinase lobes in the CDK4-cyclin D complexes caused by the elevated mobility of the N-lobe residues can weaken allosteric interactions between regulatory regions and compromise cooperativity of the inter-lobe motions that is required to trigger activating transitions. Network modelling and percolation analysis were used to emulate thermal unfolding and perform decomposition of rigid and flexible regions in the CDK2 and CDK4 complexes. These simulations showed that the percolation phase transition in the CDK2-cyclin A complexes is highly cooperative and driven by allosteric coupling between functional regions from both kinase lobes. In contrast, the imbalances in the distribution of rigid and flexible regions for the CDK4-cyclin D complexes, which are manifested by the intrinsic instability of the N-lobe, may weaken allosteric interactions and preclude productive activation. The results of this integrative computational study offer a simple and robust network-based model that explains regulatory divergences between CDK2 and CDK4 kinases.
中文翻译:

基于网络的CDK2和CDK4蛋白构象动力学和激活的建模和渗滤分析:激酶叶的动态极化和高能极化可能决定调节机制的差异
描绘细胞周期蛋白依赖性激酶(CDKs)激活机制差异基础的分子原理的总体目标对于理解紧密相关激酶之间的调节差异非常重要,这种差异可用于靶向和变构抑制剂的药物发现。为了系统地表征激活机制的动态,能量和网络特征,我们将原子模拟和弹性网络建模与残基相互作用网络的分析以及CDK2-cyclin A和CDK4-cyclin D1 / D3配合物的刚性分解相结合。这项研究的结果表明,CDK2和CDK4激活机制的差异可能是由调节区域的稳定性和变构协同作用的差异决定的。我们表明,CDK4-细胞周期蛋白D复合物中激酶裂片的差异性稳定化是由N裂片残基的增加的迁移性引起的,可以减弱调节区域之间的变构相互作用,并损害触发激活过渡所需的裂片间运动的协同作用。使用网络建模和渗滤分析来模拟热展开并执行CDK2和CDK4配合物中刚性和柔性区域的分解。这些模拟表明,CDK2-cyclin A复合物中的渗透相转变是高度协作的,并且受两个激酶叶的功能区之间的变构偶联驱动。相反,CDK4-cyclin D复合物的刚性和柔性区域分布不平衡,N瓣固有的不稳定性所表现出的“三聚体”可能会削弱变构作用并阻碍生产性激活。这项综合计算研究的结果提供了一个简单而强大的基于网络的模型,该模型解释了CDK2和CDK4激酶之间的调节差异。
更新日期:2017-10-25
中文翻译:

基于网络的CDK2和CDK4蛋白构象动力学和激活的建模和渗滤分析:激酶叶的动态极化和高能极化可能决定调节机制的差异
描绘细胞周期蛋白依赖性激酶(CDKs)激活机制差异基础的分子原理的总体目标对于理解紧密相关激酶之间的调节差异非常重要,这种差异可用于靶向和变构抑制剂的药物发现。为了系统地表征激活机制的动态,能量和网络特征,我们将原子模拟和弹性网络建模与残基相互作用网络的分析以及CDK2-cyclin A和CDK4-cyclin D1 / D3配合物的刚性分解相结合。这项研究的结果表明,CDK2和CDK4激活机制的差异可能是由调节区域的稳定性和变构协同作用的差异决定的。我们表明,CDK4-细胞周期蛋白D复合物中激酶裂片的差异性稳定化是由N裂片残基的增加的迁移性引起的,可以减弱调节区域之间的变构相互作用,并损害触发激活过渡所需的裂片间运动的协同作用。使用网络建模和渗滤分析来模拟热展开并执行CDK2和CDK4配合物中刚性和柔性区域的分解。这些模拟表明,CDK2-cyclin A复合物中的渗透相转变是高度协作的,并且受两个激酶叶的功能区之间的变构偶联驱动。相反,CDK4-cyclin D复合物的刚性和柔性区域分布不平衡,N瓣固有的不稳定性所表现出的“三聚体”可能会削弱变构作用并阻碍生产性激活。这项综合计算研究的结果提供了一个简单而强大的基于网络的模型,该模型解释了CDK2和CDK4激酶之间的调节差异。



























 京公网安备 11010802027423号
京公网安备 11010802027423号