当前位置:
X-MOL 学术
›
ACS Cent. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Computational Model and Dynamics of Monomeric Full-Length APOBEC3G
ACS Central Science ( IF 12.7 ) Pub Date : 2017-10-20 00:00:00 , DOI: 10.1021/acscentsci.7b00346 Suresh Gorle 1 , Yangang Pan 2 , Zhiqiang Sun 2 , Luda S. Shlyakhtenko 2 , Reuben S. Harris 3, 4 , Yuri L. Lyubchenko 2 , Lela Vuković 1
ACS Central Science ( IF 12.7 ) Pub Date : 2017-10-20 00:00:00 , DOI: 10.1021/acscentsci.7b00346 Suresh Gorle 1 , Yangang Pan 2 , Zhiqiang Sun 2 , Luda S. Shlyakhtenko 2 , Reuben S. Harris 3, 4 , Yuri L. Lyubchenko 2 , Lela Vuković 1
Affiliation
|
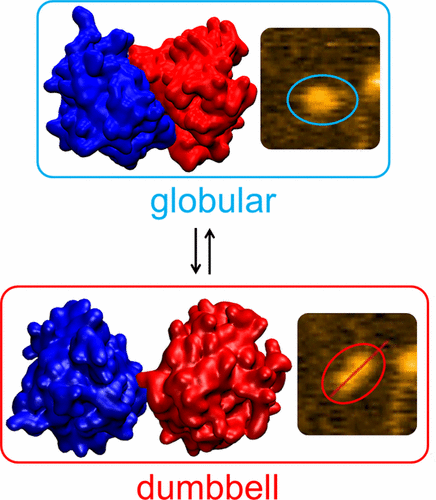
APOBEC3G (A3G) is a restriction factor that provides innate immunity against HIV-1 in the absence of viral infectivity factor (Vif) protein. However, structural information about A3G, which can aid in unraveling the mechanisms that govern its interactions and define its antiviral activity, remains unknown. Here, we built a computer model of a full-length A3G using docking approaches and molecular dynamics simulations, based on the available X-ray and NMR structural data for the two protein domains. The model revealed a large-scale dynamics of the A3G monomer, as the two A3G domains can assume compact forms or extended dumbbell type forms with domains visibly separated from each other. To validate the A3G model, we performed time-lapse high-speed atomic force microscopy (HS-AFM) experiments enabling us to get images of a fully hydrated A3G and to directly visualize its dynamics. HS-AFM confirmed that A3G exists in two forms, a globular form (∼84% of the time) and a dumbbell form (∼16% of the time), and can dynamically switch from one form to the other. The obtained HS-AFM results are in line with the computer modeling, which demonstrates a similar distribution between two forms. Furthermore, our simulations capture the complete process of A3G switching from the DNA-bound state to the closed state. The revealed dynamic nature of monomeric A3G could aid in target recognition including scanning for cytosine locations along the DNA strand and in interactions with viral RNA during packaging into HIV-1 particles.
中文翻译:

单体全长APOBEC3G的计算模型和动力学
APOBEC3G(A3G)是一种限制因子,可在缺乏病毒感染因子(Vif)蛋白的情况下提供针对HIV-1的先天免疫力。但是,有关A3G的结构性信息(仍有助于弄清控制其相互作用并定义其抗病毒活性的机制)仍然未知。在此,我们基于对接方法和分子动力学模拟,基于两个蛋白质域的可用X射线和NMR结构数据,构建了全长A3G的计算机模型。该模型揭示了A3G单体的大规模动力学,因为两个A3G域可以呈现紧凑形式或扩展的哑铃型形式,其中各个域彼此明显分开。要验证A3G模型,我们进行了延时高速原子力显微镜(HS-AFM)实验,使我们能够获取完全水合的A3G的图像并直接可视化其动力学。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。
更新日期:2017-10-20
中文翻译:

单体全长APOBEC3G的计算模型和动力学
APOBEC3G(A3G)是一种限制因子,可在缺乏病毒感染因子(Vif)蛋白的情况下提供针对HIV-1的先天免疫力。但是,有关A3G的结构性信息(仍有助于弄清控制其相互作用并定义其抗病毒活性的机制)仍然未知。在此,我们基于对接方法和分子动力学模拟,基于两个蛋白质域的可用X射线和NMR结构数据,构建了全长A3G的计算机模型。该模型揭示了A3G单体的大规模动力学,因为两个A3G域可以呈现紧凑形式或扩展的哑铃型形式,其中各个域彼此明显分开。要验证A3G模型,我们进行了延时高速原子力显微镜(HS-AFM)实验,使我们能够获取完全水合的A3G的图像并直接可视化其动力学。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。HS-AFM证实A3G有两种形式,即球形(约84%的时间)和哑铃形(约16%的时间),并且可以动态地从一种形式转换为另一种形式。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。所获得的HS-AFM结果与计算机建模相符,这表明两种形式之间的分布相似。此外,我们的仿真捕获了A3G从DNA结合状态转换为闭合状态的完整过程。单体A3G的动态特性可以帮助目标识别,包括扫描沿DNA链的胞嘧啶位置,以及在包装成HIV-1颗粒的过程中与病毒RNA的相互作用。









































 京公网安备 11010802027423号
京公网安备 11010802027423号