Virology ( IF 2.8 ) Pub Date : 2017-07-11 , DOI: 10.1016/j.virol.2017.06.026 İkbal Agah İnce , Gorben P. Pijlman , Just M. Vlak , Monique M. van Oers
|
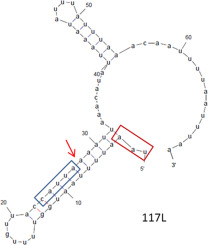
Previously, we observed that the transcripts of Invertebrate iridescent virus 6 (IIV6) are not polyadenylated, in line with the absence of canonical poly(A) motifs (AATAAA) downstream of the open reading frames (ORFs) in the genome. Here, we determined the 3′ ends of the transcripts of fifty-four IIV6 virion protein genes in infected Drosophila Schneider 2 (S2) cells. By using ligation-based amplification of cDNA ends (LACE) it was shown that the IIV6 mRNAs often ended with a CAUUA motif. In silico analysis showed that the 3′-untranslated regions of IIV6 genes have the ability to form hairpin structures (22–56 nt in length) and that for about half of all IIV6 genes these 3′ sequences contained complementary TAATG and CATTA motifs. We also show that a hairpin in the 3′ flanking region with conserved sequence motifs is a conserved feature in invertebrate-infecting iridoviruses (genus Iridovirus and Chloriridovirus).
中文翻译:

具有保守序列基序的发夹结构决定了非多腺苷酸化无脊椎动物虹膜病毒转录本的3'末端
以前,我们观察到无脊椎动物虹彩病毒6(IIV6)的转录本未聚腺苷酸化,这与基因组中开放阅读框(ORF)下游不存在规范的poly(A)模体(AATAAA)相符。在这里,我们确定了感染的果蝇施奈德2(S2)细胞中的54个IIV6病毒粒子蛋白基因的转录本的3'端。通过使用基于连接的cDNA末端扩增(LACE),显示IIV6 mRNA通常以CAUUA基序结束。在计算机上分析显示IIV6基因的3'非翻译区具有形成发夹结构的能力(长度为22-56 nt),并且对于所有IIV6基因的大约一半,这些3'序列均包含互补的TAATG和CATTA基序。我们还显示,在3'侧翼区域具有保守序列基序的发夹是无脊椎动物感染的虹膜病毒(虹膜病毒和绿藻病毒)的保守特征。











































 京公网安备 11010802027423号
京公网安备 11010802027423号