当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Chemical Proteomics Strategies for Analyzing Protein Lipidation Reveal the Bacterial O-Mycoloylome
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2024-04-18 , DOI: 10.1021/jacs.4c02278 Nicholas Banahene 1, 2 , Trenton M. Peters-Clarke 3, 4, 5 , Kyle J. Biegas 1, 2 , Evgenia Shishkova 4, 5 , Elizabeth M. Hart 6, 7 , Amelia C. McKitterick 6, 7 , Nikolas H. Kambitsis 1 , Ulysses G. Johnson 1, 2 , Thomas G. Bernhardt 6, 7 , Joshua J. Coon 3, 4, 5, 8 , Benjamin M. Swarts 1, 2
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2024-04-18 , DOI: 10.1021/jacs.4c02278 Nicholas Banahene 1, 2 , Trenton M. Peters-Clarke 3, 4, 5 , Kyle J. Biegas 1, 2 , Evgenia Shishkova 4, 5 , Elizabeth M. Hart 6, 7 , Amelia C. McKitterick 6, 7 , Nikolas H. Kambitsis 1 , Ulysses G. Johnson 1, 2 , Thomas G. Bernhardt 6, 7 , Joshua J. Coon 3, 4, 5, 8 , Benjamin M. Swarts 1, 2
Affiliation
|
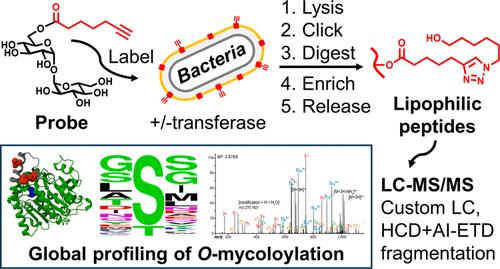
Protein lipidation dynamically controls protein localization and function within cellular membranes. A unique form of protein O-fatty acylation in Corynebacterium, termed protein O-mycoloylation, involves the attachment of mycolic acids─unusually large and hydrophobic fatty acids─to serine residues of proteins in these organisms’ outer mycomembrane. However, as with other forms of protein lipidation, the scope and functional consequences of protein O-mycoloylation are challenging to investigate due to the inherent difficulties of enriching and analyzing lipidated peptides. To facilitate the analysis of protein lipidation and enable the comprehensive profiling and site mapping of protein O-mycoloylation, we developed a chemical proteomics strategy integrating metabolic labeling, click chemistry, cleavable linkers, and a novel liquid chromatography-tandem mass spectrometry (LC-MS/MS) method employing LC separation and complementary fragmentation methods tailored to the analysis of lipophilic, MS-labile O-acylated peptides. Using these tools in the model organism Corynebacterium glutamicum, we identified approximately 30 candidate O-mycoloylated proteins, including porins, mycoloyltransferases, secreted hydrolases, and other proteins with cell envelope-related functions─consistent with a role for O-mycoloylation in targeting proteins to the mycomembrane. Site mapping revealed that many of the proteins contained multiple spatially proximal modification sites, which occurred predominantly at serine residues surrounded by conformationally flexible peptide motifs. Overall, this study (i) discloses the putative protein O-mycoloylome for the first time, (ii) yields new insights into the undercharacterized proteome of the mycomembrane, which is a hallmark of important pathogens (e.g., Corynebacterium diphtheriae, Mycobacterium tuberculosis), and (iii) provides generally applicable chemical strategies for the proteomic analysis of protein lipidation.
中文翻译:

分析蛋白质脂质化的化学蛋白质组学策略揭示细菌 O-Mycoloylome
蛋白质脂化动态控制蛋白质在细胞膜内的定位和功能。棒状杆菌中蛋白质O - 脂肪酰化的一种独特形式,称为蛋白质O - 菌酰化,涉及将分枝菌酸(异常大且疏水的脂肪酸)附着到这些生物体外菌膜中蛋白质的丝氨酸残基上。然而,与其他形式的蛋白质脂化一样,由于富集和分析脂化肽的固有困难,蛋白质O-菌酰化的范围和功能后果的研究具有挑战性。为了促进蛋白质脂化分析并实现蛋白质O菌酰化的全面分析和位点定位,我们开发了一种化学蛋白质组学策略,集成了代谢标记、点击化学、可裂解接头和新型液相色谱-串联质谱法 (LC-MS) /MS) 方法采用 LC 分离和互补片段化方法,专为分析亲脂性、MS 不稳定的O-酰化肽而设计。在模式生物谷氨酸棒杆菌中使用这些工具,我们鉴定了大约 30 种候选O - 菌酰化蛋白,包括孔蛋白、菌酰转移酶、分泌性水解酶和其他具有细胞包膜相关功能的蛋白质,这与O - 菌酰化在靶向蛋白质中的作用一致。菌膜。位点图谱显示,许多蛋白质含有多个空间上邻近的修饰位点,这些修饰位点主要发生在被构象灵活的肽基序包围的丝氨酸残基处。总体而言,这项研究(i)首次公开了假定的蛋白质O -mycoloylome,(ii)对菌膜蛋白质组的表征不足产生了新的见解,这是重要病原体(例如白喉棒杆菌、结核分枝杆菌)的标志, (iii) 为蛋白质脂化的蛋白质组学分析提供普遍适用的化学策略。
更新日期:2024-04-18
中文翻译:

分析蛋白质脂质化的化学蛋白质组学策略揭示细菌 O-Mycoloylome
蛋白质脂化动态控制蛋白质在细胞膜内的定位和功能。棒状杆菌中蛋白质O - 脂肪酰化的一种独特形式,称为蛋白质O - 菌酰化,涉及将分枝菌酸(异常大且疏水的脂肪酸)附着到这些生物体外菌膜中蛋白质的丝氨酸残基上。然而,与其他形式的蛋白质脂化一样,由于富集和分析脂化肽的固有困难,蛋白质O-菌酰化的范围和功能后果的研究具有挑战性。为了促进蛋白质脂化分析并实现蛋白质O菌酰化的全面分析和位点定位,我们开发了一种化学蛋白质组学策略,集成了代谢标记、点击化学、可裂解接头和新型液相色谱-串联质谱法 (LC-MS) /MS) 方法采用 LC 分离和互补片段化方法,专为分析亲脂性、MS 不稳定的O-酰化肽而设计。在模式生物谷氨酸棒杆菌中使用这些工具,我们鉴定了大约 30 种候选O - 菌酰化蛋白,包括孔蛋白、菌酰转移酶、分泌性水解酶和其他具有细胞包膜相关功能的蛋白质,这与O - 菌酰化在靶向蛋白质中的作用一致。菌膜。位点图谱显示,许多蛋白质含有多个空间上邻近的修饰位点,这些修饰位点主要发生在被构象灵活的肽基序包围的丝氨酸残基处。总体而言,这项研究(i)首次公开了假定的蛋白质O -mycoloylome,(ii)对菌膜蛋白质组的表征不足产生了新的见解,这是重要病原体(例如白喉棒杆菌、结核分枝杆菌)的标志, (iii) 为蛋白质脂化的蛋白质组学分析提供普遍适用的化学策略。



























 京公网安备 11010802027423号
京公网安备 11010802027423号